
Pteropus associated gemycircularvirus 10
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ptero10
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
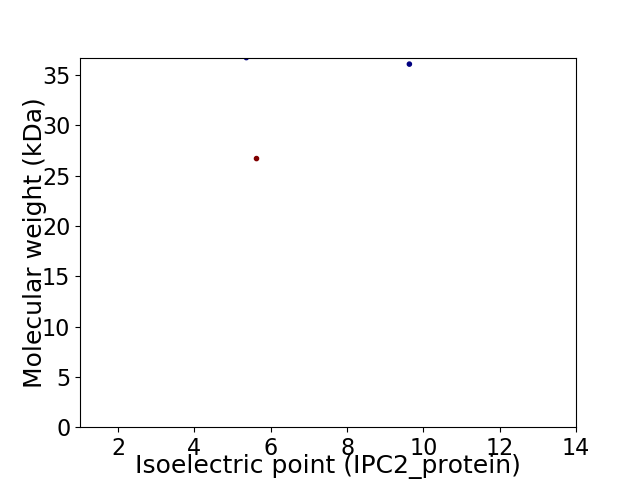
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
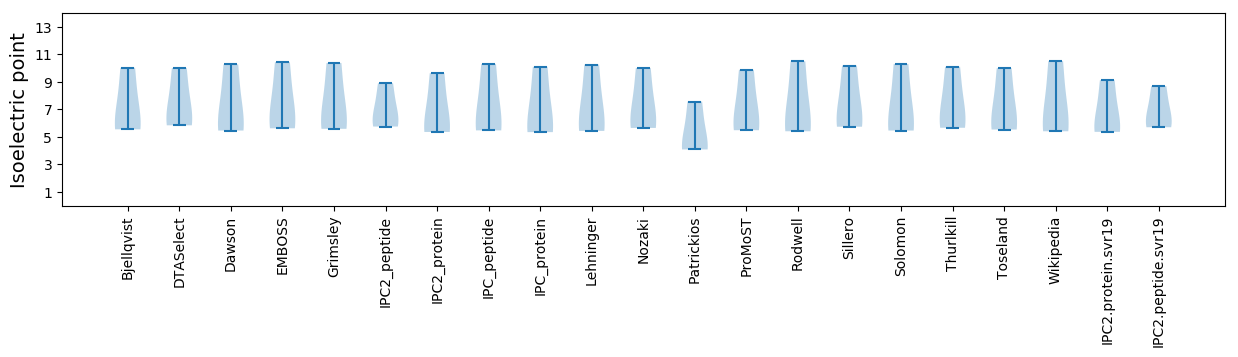
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTL6|A0A140CTL6_9VIRU RepA OS=Pteropus associated gemycircularvirus 10 OX=1985404 PE=3 SV=1
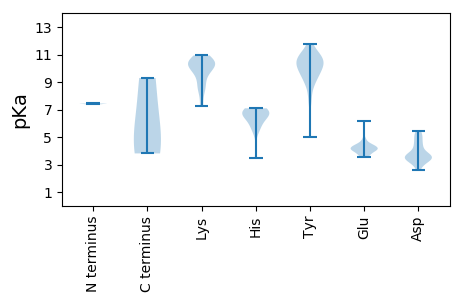
MM1 pKa = 7.44TFHH4 pKa = 7.09FSARR8 pKa = 11.84YY9 pKa = 9.48VLLTYY14 pKa = 10.06AQSGDD19 pKa = 3.41LSEE22 pKa = 4.41WSVLDD27 pKa = 4.62HH28 pKa = 6.79ISSLGAEE35 pKa = 4.41CIVARR40 pKa = 11.84EE41 pKa = 4.04DD42 pKa = 3.46HH43 pKa = 6.99ADD45 pKa = 3.7GGTHH49 pKa = 6.54LHH51 pKa = 5.76VFCDD55 pKa = 5.07FGRR58 pKa = 11.84KK59 pKa = 8.32KK60 pKa = 10.15QSRR63 pKa = 11.84RR64 pKa = 11.84PDD66 pKa = 3.36YY67 pKa = 10.83FDD69 pKa = 4.99VEE71 pKa = 4.4GHH73 pKa = 6.6HH74 pKa = 6.71PNIVPSRR81 pKa = 11.84GRR83 pKa = 11.84AGEE86 pKa = 4.02GWDD89 pKa = 3.56YY90 pKa = 10.72ATKK93 pKa = 10.53DD94 pKa = 3.54GNIVAGGLEE103 pKa = 4.1RR104 pKa = 11.84PGGGGLPSTPDD115 pKa = 2.6KK116 pKa = 10.39WRR118 pKa = 11.84EE119 pKa = 3.61IVGAEE124 pKa = 3.82SRR126 pKa = 11.84EE127 pKa = 3.99EE128 pKa = 4.19FFDD131 pKa = 5.34LLRR134 pKa = 11.84QLDD137 pKa = 4.38PKK139 pKa = 10.33TLITRR144 pKa = 11.84WSEE147 pKa = 3.6LNRR150 pKa = 11.84YY151 pKa = 9.13ADD153 pKa = 3.12AAYY156 pKa = 7.63ATRR159 pKa = 11.84AEE161 pKa = 4.79PYY163 pKa = 9.97VGPTGIEE170 pKa = 4.16FEE172 pKa = 4.44LGMVPEE178 pKa = 4.37LARR181 pKa = 11.84WGRR184 pKa = 11.84EE185 pKa = 3.75LPVLYY190 pKa = 10.58GPSRR194 pKa = 11.84LGKK197 pKa = 7.27TLWARR202 pKa = 11.84SLGPHH207 pKa = 6.96AYY209 pKa = 9.55IMGMVSGHH217 pKa = 5.28VLTRR221 pKa = 11.84DD222 pKa = 3.44MPDD225 pKa = 2.6AQYY228 pKa = 11.73AVFDD232 pKa = 4.43DD233 pKa = 3.82MRR235 pKa = 11.84GGISMFPSFKK245 pKa = 10.24EE246 pKa = 3.57WFGAQEE252 pKa = 4.17VVTVKK257 pKa = 9.72TLYY260 pKa = 10.24RR261 pKa = 11.84DD262 pKa = 3.61PVQMKK267 pKa = 8.97WGKK270 pKa = 8.84PCIWLANADD279 pKa = 3.96PRR281 pKa = 11.84DD282 pKa = 3.7QLKK285 pKa = 10.93ADD287 pKa = 3.28ITDD290 pKa = 3.66RR291 pKa = 11.84TPKK294 pKa = 10.53GRR296 pKa = 11.84IDD298 pKa = 5.41LIYY301 pKa = 10.94NDD303 pKa = 4.47IEE305 pKa = 4.1WMEE308 pKa = 4.19ANCIFVGLQEE318 pKa = 4.88AIFRR322 pKa = 11.84ASTEE326 pKa = 3.81
MM1 pKa = 7.44TFHH4 pKa = 7.09FSARR8 pKa = 11.84YY9 pKa = 9.48VLLTYY14 pKa = 10.06AQSGDD19 pKa = 3.41LSEE22 pKa = 4.41WSVLDD27 pKa = 4.62HH28 pKa = 6.79ISSLGAEE35 pKa = 4.41CIVARR40 pKa = 11.84EE41 pKa = 4.04DD42 pKa = 3.46HH43 pKa = 6.99ADD45 pKa = 3.7GGTHH49 pKa = 6.54LHH51 pKa = 5.76VFCDD55 pKa = 5.07FGRR58 pKa = 11.84KK59 pKa = 8.32KK60 pKa = 10.15QSRR63 pKa = 11.84RR64 pKa = 11.84PDD66 pKa = 3.36YY67 pKa = 10.83FDD69 pKa = 4.99VEE71 pKa = 4.4GHH73 pKa = 6.6HH74 pKa = 6.71PNIVPSRR81 pKa = 11.84GRR83 pKa = 11.84AGEE86 pKa = 4.02GWDD89 pKa = 3.56YY90 pKa = 10.72ATKK93 pKa = 10.53DD94 pKa = 3.54GNIVAGGLEE103 pKa = 4.1RR104 pKa = 11.84PGGGGLPSTPDD115 pKa = 2.6KK116 pKa = 10.39WRR118 pKa = 11.84EE119 pKa = 3.61IVGAEE124 pKa = 3.82SRR126 pKa = 11.84EE127 pKa = 3.99EE128 pKa = 4.19FFDD131 pKa = 5.34LLRR134 pKa = 11.84QLDD137 pKa = 4.38PKK139 pKa = 10.33TLITRR144 pKa = 11.84WSEE147 pKa = 3.6LNRR150 pKa = 11.84YY151 pKa = 9.13ADD153 pKa = 3.12AAYY156 pKa = 7.63ATRR159 pKa = 11.84AEE161 pKa = 4.79PYY163 pKa = 9.97VGPTGIEE170 pKa = 4.16FEE172 pKa = 4.44LGMVPEE178 pKa = 4.37LARR181 pKa = 11.84WGRR184 pKa = 11.84EE185 pKa = 3.75LPVLYY190 pKa = 10.58GPSRR194 pKa = 11.84LGKK197 pKa = 7.27TLWARR202 pKa = 11.84SLGPHH207 pKa = 6.96AYY209 pKa = 9.55IMGMVSGHH217 pKa = 5.28VLTRR221 pKa = 11.84DD222 pKa = 3.44MPDD225 pKa = 2.6AQYY228 pKa = 11.73AVFDD232 pKa = 4.43DD233 pKa = 3.82MRR235 pKa = 11.84GGISMFPSFKK245 pKa = 10.24EE246 pKa = 3.57WFGAQEE252 pKa = 4.17VVTVKK257 pKa = 9.72TLYY260 pKa = 10.24RR261 pKa = 11.84DD262 pKa = 3.61PVQMKK267 pKa = 8.97WGKK270 pKa = 8.84PCIWLANADD279 pKa = 3.96PRR281 pKa = 11.84DD282 pKa = 3.7QLKK285 pKa = 10.93ADD287 pKa = 3.28ITDD290 pKa = 3.66RR291 pKa = 11.84TPKK294 pKa = 10.53GRR296 pKa = 11.84IDD298 pKa = 5.41LIYY301 pKa = 10.94NDD303 pKa = 4.47IEE305 pKa = 4.1WMEE308 pKa = 4.19ANCIFVGLQEE318 pKa = 4.88AIFRR322 pKa = 11.84ASTEE326 pKa = 3.81
Molecular weight: 36.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTL5|A0A140CTL5_9VIRU Replication-associated protein OS=Pteropus associated gemycircularvirus 10 OX=1985404 PE=3 SV=1
MM1 pKa = 7.38TLHH4 pKa = 6.55SLAARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 9.59AKK14 pKa = 10.66LMASRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VPAGRR27 pKa = 11.84RR28 pKa = 11.84SRR30 pKa = 11.84PRR32 pKa = 11.84SRR34 pKa = 11.84YY35 pKa = 5.02TAKK38 pKa = 9.56TRR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 8.55APKK45 pKa = 10.12RR46 pKa = 11.84PMSKK50 pKa = 10.14KK51 pKa = 10.3RR52 pKa = 11.84ILDD55 pKa = 3.21VTTVKK60 pKa = 10.51KK61 pKa = 10.16RR62 pKa = 11.84DD63 pKa = 3.51NMPCYY68 pKa = 10.68TNMTNSGAGTTFSITAAYY86 pKa = 8.85IRR88 pKa = 11.84PNTSSDD94 pKa = 3.39PLINHH99 pKa = 5.99PTVMLWNATARR110 pKa = 11.84DD111 pKa = 3.98LTNSAGTINIRR122 pKa = 11.84AVEE125 pKa = 4.16SARR128 pKa = 11.84TSSTPYY134 pKa = 9.78MVGLKK139 pKa = 8.84EE140 pKa = 4.09TVEE143 pKa = 4.28IQASSGMPWQWRR155 pKa = 11.84RR156 pKa = 11.84ICFTYY161 pKa = 10.34KK162 pKa = 10.72GILPGSTPTTTYY174 pKa = 9.73YY175 pKa = 10.25PYY177 pKa = 10.58RR178 pKa = 11.84EE179 pKa = 4.08TSSGFVRR186 pKa = 11.84VASPLSGDD194 pKa = 3.65RR195 pKa = 11.84NSGATYY201 pKa = 10.99DD202 pKa = 3.68LFYY205 pKa = 10.98LIFAGQNASDD215 pKa = 3.47WTDD218 pKa = 3.33PMVAKK223 pKa = 10.57LDD225 pKa = 3.52QTRR228 pKa = 11.84ISVKK232 pKa = 9.58YY233 pKa = 10.28DD234 pKa = 2.92KK235 pKa = 10.89TRR237 pKa = 11.84TLSCGNDD244 pKa = 3.26DD245 pKa = 4.37GSIRR249 pKa = 11.84KK250 pKa = 8.5YY251 pKa = 10.53KK252 pKa = 9.66HH253 pKa = 3.45WHH255 pKa = 5.77AMGKK259 pKa = 7.26TLAYY263 pKa = 10.47DD264 pKa = 3.78DD265 pKa = 5.27DD266 pKa = 4.91EE267 pKa = 6.14NGEE270 pKa = 4.14NMITNYY276 pKa = 10.25QSVDD280 pKa = 3.16SRR282 pKa = 11.84VGMGDD287 pKa = 3.72YY288 pKa = 10.87YY289 pKa = 11.55VVDD292 pKa = 4.2LFRR295 pKa = 11.84SRR297 pKa = 11.84FGATASDD304 pKa = 3.46SLLFNPAATLYY315 pKa = 9.05WHH317 pKa = 7.05EE318 pKa = 4.35KK319 pKa = 9.3
MM1 pKa = 7.38TLHH4 pKa = 6.55SLAARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 9.59AKK14 pKa = 10.66LMASRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VPAGRR27 pKa = 11.84RR28 pKa = 11.84SRR30 pKa = 11.84PRR32 pKa = 11.84SRR34 pKa = 11.84YY35 pKa = 5.02TAKK38 pKa = 9.56TRR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 8.55APKK45 pKa = 10.12RR46 pKa = 11.84PMSKK50 pKa = 10.14KK51 pKa = 10.3RR52 pKa = 11.84ILDD55 pKa = 3.21VTTVKK60 pKa = 10.51KK61 pKa = 10.16RR62 pKa = 11.84DD63 pKa = 3.51NMPCYY68 pKa = 10.68TNMTNSGAGTTFSITAAYY86 pKa = 8.85IRR88 pKa = 11.84PNTSSDD94 pKa = 3.39PLINHH99 pKa = 5.99PTVMLWNATARR110 pKa = 11.84DD111 pKa = 3.98LTNSAGTINIRR122 pKa = 11.84AVEE125 pKa = 4.16SARR128 pKa = 11.84TSSTPYY134 pKa = 9.78MVGLKK139 pKa = 8.84EE140 pKa = 4.09TVEE143 pKa = 4.28IQASSGMPWQWRR155 pKa = 11.84RR156 pKa = 11.84ICFTYY161 pKa = 10.34KK162 pKa = 10.72GILPGSTPTTTYY174 pKa = 9.73YY175 pKa = 10.25PYY177 pKa = 10.58RR178 pKa = 11.84EE179 pKa = 4.08TSSGFVRR186 pKa = 11.84VASPLSGDD194 pKa = 3.65RR195 pKa = 11.84NSGATYY201 pKa = 10.99DD202 pKa = 3.68LFYY205 pKa = 10.98LIFAGQNASDD215 pKa = 3.47WTDD218 pKa = 3.33PMVAKK223 pKa = 10.57LDD225 pKa = 3.52QTRR228 pKa = 11.84ISVKK232 pKa = 9.58YY233 pKa = 10.28DD234 pKa = 2.92KK235 pKa = 10.89TRR237 pKa = 11.84TLSCGNDD244 pKa = 3.26DD245 pKa = 4.37GSIRR249 pKa = 11.84KK250 pKa = 8.5YY251 pKa = 10.53KK252 pKa = 9.66HH253 pKa = 3.45WHH255 pKa = 5.77AMGKK259 pKa = 7.26TLAYY263 pKa = 10.47DD264 pKa = 3.78DD265 pKa = 5.27DD266 pKa = 4.91EE267 pKa = 6.14NGEE270 pKa = 4.14NMITNYY276 pKa = 10.25QSVDD280 pKa = 3.16SRR282 pKa = 11.84VGMGDD287 pKa = 3.72YY288 pKa = 10.87YY289 pKa = 11.55VVDD292 pKa = 4.2LFRR295 pKa = 11.84SRR297 pKa = 11.84FGATASDD304 pKa = 3.46SLLFNPAATLYY315 pKa = 9.05WHH317 pKa = 7.05EE318 pKa = 4.35KK319 pKa = 9.3
Molecular weight: 36.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
886 |
241 |
326 |
295.3 |
33.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.126 ± 0.139 | 1.354 ± 0.263 |
6.433 ± 0.423 | 5.192 ± 1.225 |
3.612 ± 0.453 | 8.804 ± 1.206 |
2.483 ± 0.396 | 4.176 ± 0.385 |
3.837 ± 0.557 | 7.675 ± 0.793 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.822 ± 0.464 | 3.047 ± 0.695 |
5.418 ± 0.196 | 1.919 ± 0.24 |
8.691 ± 0.57 | 7.111 ± 1.001 |
6.772 ± 1.46 | 5.643 ± 0.429 |
2.483 ± 0.284 | 4.402 ± 0.64 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
