
Hubei tombus-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
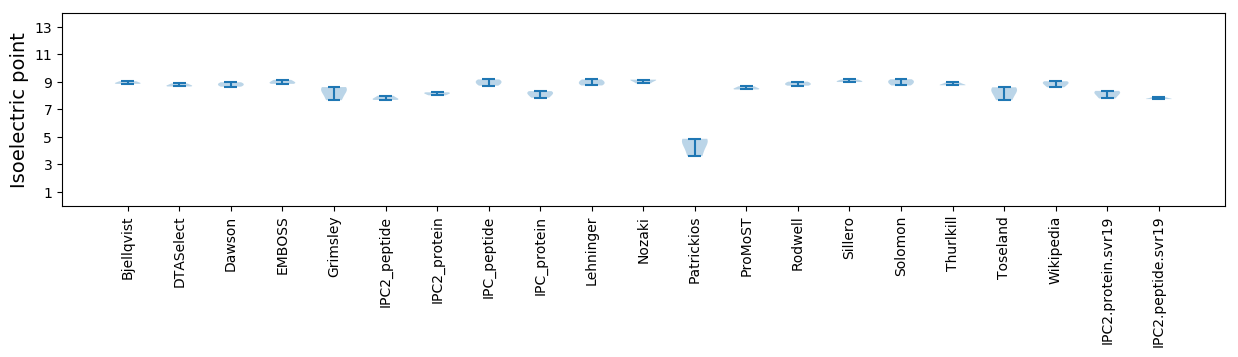
Average proteome isoelectric point is 8.11
Get precalculated fractions of proteins
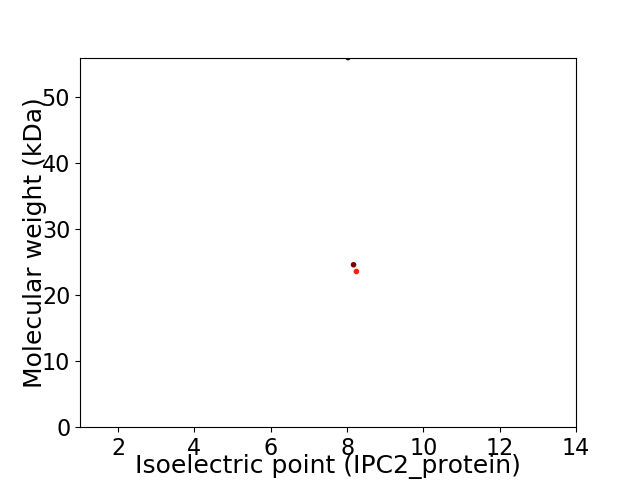
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGE9|A0A1L3KGE9_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 4 OX=1923288 PE=4 SV=1
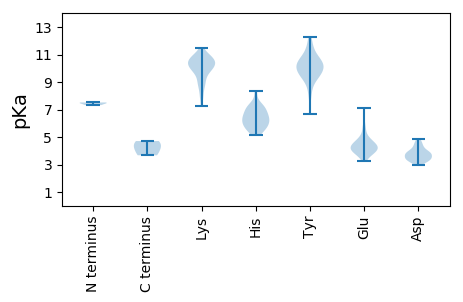
MM1 pKa = 7.51GKK3 pKa = 9.25QRR5 pKa = 11.84AVTQIGRR12 pKa = 11.84VAPPNRR18 pKa = 11.84AAVHH22 pKa = 6.53DD23 pKa = 4.31NSVLNCDD30 pKa = 2.97RR31 pKa = 11.84ALRR34 pKa = 11.84EE35 pKa = 3.28RR36 pKa = 11.84VYY38 pKa = 10.76YY39 pKa = 10.12VKK41 pKa = 10.97NGDD44 pKa = 3.4GFARR48 pKa = 11.84PPQPNPSSVLTLGQFRR64 pKa = 11.84KK65 pKa = 10.02KK66 pKa = 10.51LSRR69 pKa = 11.84CVPRR73 pKa = 11.84IPALSWEE80 pKa = 4.66LFPSRR85 pKa = 11.84YY86 pKa = 9.69KK87 pKa = 10.46DD88 pKa = 3.09AKK90 pKa = 9.95KK91 pKa = 10.73RR92 pKa = 11.84NIYY95 pKa = 10.4KK96 pKa = 10.5NADD99 pKa = 3.27QSLHH103 pKa = 5.69EE104 pKa = 4.96RR105 pKa = 11.84PLSIHH110 pKa = 6.87DD111 pKa = 4.13SYY113 pKa = 12.26VKK115 pKa = 10.79GFIKK119 pKa = 10.39DD120 pKa = 3.46EE121 pKa = 4.85KK122 pKa = 10.02INLMKK127 pKa = 10.64KK128 pKa = 9.43EE129 pKa = 4.2DD130 pKa = 3.92PVPRR134 pKa = 11.84AILPLSPRR142 pKa = 11.84LNIVEE147 pKa = 4.11GSIISHH153 pKa = 6.51RR154 pKa = 11.84EE155 pKa = 3.38HH156 pKa = 7.3AVFEE160 pKa = 4.96GIDD163 pKa = 3.43LVFGRR168 pKa = 11.84KK169 pKa = 7.25TVMKK173 pKa = 10.23GLNAEE178 pKa = 4.03EE179 pKa = 4.36KK180 pKa = 10.86GRR182 pKa = 11.84IIAEE186 pKa = 3.87KK187 pKa = 9.91CGRR190 pKa = 11.84FAKK193 pKa = 10.51FAALSIDD200 pKa = 3.35MAKK203 pKa = 10.13FDD205 pKa = 3.57QHH207 pKa = 8.33VSVLMNMFEE216 pKa = 4.16NSVLVDD222 pKa = 3.62TCCTPQEE229 pKa = 3.92RR230 pKa = 11.84RR231 pKa = 11.84EE232 pKa = 3.99LSKK235 pKa = 10.6ILAWSIHH242 pKa = 5.55SKK244 pKa = 10.93GRR246 pKa = 11.84MQATDD251 pKa = 3.18GKK253 pKa = 11.01VNFIRR258 pKa = 11.84EE259 pKa = 4.28GGRR262 pKa = 11.84LSGCMHH268 pKa = 7.07TSLGNIIIMCAMFWTFMQQFGDD290 pKa = 3.93IKK292 pKa = 10.12WDD294 pKa = 3.91YY295 pKa = 11.37INDD298 pKa = 3.68GDD300 pKa = 4.22DD301 pKa = 3.21CVLFVEE307 pKa = 5.79DD308 pKa = 4.84KK309 pKa = 10.65DD310 pKa = 3.87LHH312 pKa = 6.17KK313 pKa = 10.66VAPKK317 pKa = 9.89IQDD320 pKa = 3.3YY321 pKa = 9.86FLGMGFTAVVEE332 pKa = 4.41DD333 pKa = 4.03VVHH336 pKa = 7.27DD337 pKa = 3.51IEE339 pKa = 7.11KK340 pKa = 10.11IDD342 pKa = 3.88FCQSRR347 pKa = 11.84PVWDD351 pKa = 3.96GDD353 pKa = 3.9DD354 pKa = 4.81YY355 pKa = 11.77IMCRR359 pKa = 11.84NPHH362 pKa = 5.4TALIKK367 pKa = 10.24DD368 pKa = 4.07TMCLKK373 pKa = 10.73PIGNEE378 pKa = 4.03NEE380 pKa = 3.58WGSWVRR386 pKa = 11.84SVSEE390 pKa = 3.81SGIAGFAGMPIFQSFYY406 pKa = 9.21EE407 pKa = 4.32LYY409 pKa = 10.3SRR411 pKa = 11.84SSKK414 pKa = 10.28GFKK417 pKa = 9.74VNKK420 pKa = 8.9LHH422 pKa = 7.38RR423 pKa = 11.84VDD425 pKa = 4.45GGLKK429 pKa = 8.67IATRR433 pKa = 11.84GMHH436 pKa = 6.02RR437 pKa = 11.84KK438 pKa = 8.97SRR440 pKa = 11.84PIDD443 pKa = 3.38DD444 pKa = 3.53RR445 pKa = 11.84TRR447 pKa = 11.84YY448 pKa = 10.19SFWLAWGVLPDD459 pKa = 3.76AQICTEE465 pKa = 4.0EE466 pKa = 4.48MFSTMHH472 pKa = 6.71LNYY475 pKa = 9.4HH476 pKa = 6.53DD477 pKa = 4.56PVLSFEE483 pKa = 4.43SLSLPSYY490 pKa = 10.25FF491 pKa = 4.71
MM1 pKa = 7.51GKK3 pKa = 9.25QRR5 pKa = 11.84AVTQIGRR12 pKa = 11.84VAPPNRR18 pKa = 11.84AAVHH22 pKa = 6.53DD23 pKa = 4.31NSVLNCDD30 pKa = 2.97RR31 pKa = 11.84ALRR34 pKa = 11.84EE35 pKa = 3.28RR36 pKa = 11.84VYY38 pKa = 10.76YY39 pKa = 10.12VKK41 pKa = 10.97NGDD44 pKa = 3.4GFARR48 pKa = 11.84PPQPNPSSVLTLGQFRR64 pKa = 11.84KK65 pKa = 10.02KK66 pKa = 10.51LSRR69 pKa = 11.84CVPRR73 pKa = 11.84IPALSWEE80 pKa = 4.66LFPSRR85 pKa = 11.84YY86 pKa = 9.69KK87 pKa = 10.46DD88 pKa = 3.09AKK90 pKa = 9.95KK91 pKa = 10.73RR92 pKa = 11.84NIYY95 pKa = 10.4KK96 pKa = 10.5NADD99 pKa = 3.27QSLHH103 pKa = 5.69EE104 pKa = 4.96RR105 pKa = 11.84PLSIHH110 pKa = 6.87DD111 pKa = 4.13SYY113 pKa = 12.26VKK115 pKa = 10.79GFIKK119 pKa = 10.39DD120 pKa = 3.46EE121 pKa = 4.85KK122 pKa = 10.02INLMKK127 pKa = 10.64KK128 pKa = 9.43EE129 pKa = 4.2DD130 pKa = 3.92PVPRR134 pKa = 11.84AILPLSPRR142 pKa = 11.84LNIVEE147 pKa = 4.11GSIISHH153 pKa = 6.51RR154 pKa = 11.84EE155 pKa = 3.38HH156 pKa = 7.3AVFEE160 pKa = 4.96GIDD163 pKa = 3.43LVFGRR168 pKa = 11.84KK169 pKa = 7.25TVMKK173 pKa = 10.23GLNAEE178 pKa = 4.03EE179 pKa = 4.36KK180 pKa = 10.86GRR182 pKa = 11.84IIAEE186 pKa = 3.87KK187 pKa = 9.91CGRR190 pKa = 11.84FAKK193 pKa = 10.51FAALSIDD200 pKa = 3.35MAKK203 pKa = 10.13FDD205 pKa = 3.57QHH207 pKa = 8.33VSVLMNMFEE216 pKa = 4.16NSVLVDD222 pKa = 3.62TCCTPQEE229 pKa = 3.92RR230 pKa = 11.84RR231 pKa = 11.84EE232 pKa = 3.99LSKK235 pKa = 10.6ILAWSIHH242 pKa = 5.55SKK244 pKa = 10.93GRR246 pKa = 11.84MQATDD251 pKa = 3.18GKK253 pKa = 11.01VNFIRR258 pKa = 11.84EE259 pKa = 4.28GGRR262 pKa = 11.84LSGCMHH268 pKa = 7.07TSLGNIIIMCAMFWTFMQQFGDD290 pKa = 3.93IKK292 pKa = 10.12WDD294 pKa = 3.91YY295 pKa = 11.37INDD298 pKa = 3.68GDD300 pKa = 4.22DD301 pKa = 3.21CVLFVEE307 pKa = 5.79DD308 pKa = 4.84KK309 pKa = 10.65DD310 pKa = 3.87LHH312 pKa = 6.17KK313 pKa = 10.66VAPKK317 pKa = 9.89IQDD320 pKa = 3.3YY321 pKa = 9.86FLGMGFTAVVEE332 pKa = 4.41DD333 pKa = 4.03VVHH336 pKa = 7.27DD337 pKa = 3.51IEE339 pKa = 7.11KK340 pKa = 10.11IDD342 pKa = 3.88FCQSRR347 pKa = 11.84PVWDD351 pKa = 3.96GDD353 pKa = 3.9DD354 pKa = 4.81YY355 pKa = 11.77IMCRR359 pKa = 11.84NPHH362 pKa = 5.4TALIKK367 pKa = 10.24DD368 pKa = 4.07TMCLKK373 pKa = 10.73PIGNEE378 pKa = 4.03NEE380 pKa = 3.58WGSWVRR386 pKa = 11.84SVSEE390 pKa = 3.81SGIAGFAGMPIFQSFYY406 pKa = 9.21EE407 pKa = 4.32LYY409 pKa = 10.3SRR411 pKa = 11.84SSKK414 pKa = 10.28GFKK417 pKa = 9.74VNKK420 pKa = 8.9LHH422 pKa = 7.38RR423 pKa = 11.84VDD425 pKa = 4.45GGLKK429 pKa = 8.67IATRR433 pKa = 11.84GMHH436 pKa = 6.02RR437 pKa = 11.84KK438 pKa = 8.97SRR440 pKa = 11.84PIDD443 pKa = 3.38DD444 pKa = 3.53RR445 pKa = 11.84TRR447 pKa = 11.84YY448 pKa = 10.19SFWLAWGVLPDD459 pKa = 3.76AQICTEE465 pKa = 4.0EE466 pKa = 4.48MFSTMHH472 pKa = 6.71LNYY475 pKa = 9.4HH476 pKa = 6.53DD477 pKa = 4.56PVLSFEE483 pKa = 4.43SLSLPSYY490 pKa = 10.25FF491 pKa = 4.71
Molecular weight: 56.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGE9|A0A1L3KGE9_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 4 OX=1923288 PE=4 SV=1
MM1 pKa = 7.47ATTLLIPAYY10 pKa = 9.56IATIHH15 pKa = 6.18AADD18 pKa = 4.27YY19 pKa = 9.55AWSVYY24 pKa = 9.26TSSTNPNDD32 pKa = 3.52DD33 pKa = 3.35SFEE36 pKa = 4.39RR37 pKa = 11.84RR38 pKa = 11.84VEE40 pKa = 3.97WNRR43 pKa = 11.84VKK45 pKa = 10.45SWFRR49 pKa = 11.84GTTALSRR56 pKa = 11.84EE57 pKa = 4.34DD58 pKa = 3.34EE59 pKa = 4.25RR60 pKa = 11.84RR61 pKa = 11.84GHH63 pKa = 5.94NMLRR67 pKa = 11.84EE68 pKa = 4.21FNSEE72 pKa = 3.91EE73 pKa = 4.37SGDD76 pKa = 3.82LPTFEE81 pKa = 6.61DD82 pKa = 3.71EE83 pKa = 5.43DD84 pKa = 4.6LFDD87 pKa = 4.58VEE89 pKa = 5.65LDD91 pKa = 3.76GEE93 pKa = 4.79GKK95 pKa = 8.1VTRR98 pKa = 11.84KK99 pKa = 9.55KK100 pKa = 9.95IRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84MHH106 pKa = 5.87KK107 pKa = 9.82PFVHH111 pKa = 6.22RR112 pKa = 11.84LVRR115 pKa = 11.84HH116 pKa = 5.65CRR118 pKa = 11.84GEE120 pKa = 3.81LGQRR124 pKa = 11.84VHH126 pKa = 6.57TPPNVMVVEE135 pKa = 4.85RR136 pKa = 11.84TARR139 pKa = 11.84AYY141 pKa = 10.46CHH143 pKa = 5.13EE144 pKa = 4.56HH145 pKa = 5.52YY146 pKa = 10.75VRR148 pKa = 11.84SSDD151 pKa = 3.2ISCILPVVVALYY163 pKa = 9.29FFSRR167 pKa = 11.84SDD169 pKa = 3.31VQIRR173 pKa = 11.84TEE175 pKa = 3.8ALKK178 pKa = 10.7QSSAFVKK185 pKa = 10.02SSKK188 pKa = 7.81PHH190 pKa = 6.0RR191 pKa = 11.84FVGVGGRR198 pKa = 11.84QHH200 pKa = 6.37GVADD204 pKa = 3.77AA205 pKa = 4.29
MM1 pKa = 7.47ATTLLIPAYY10 pKa = 9.56IATIHH15 pKa = 6.18AADD18 pKa = 4.27YY19 pKa = 9.55AWSVYY24 pKa = 9.26TSSTNPNDD32 pKa = 3.52DD33 pKa = 3.35SFEE36 pKa = 4.39RR37 pKa = 11.84RR38 pKa = 11.84VEE40 pKa = 3.97WNRR43 pKa = 11.84VKK45 pKa = 10.45SWFRR49 pKa = 11.84GTTALSRR56 pKa = 11.84EE57 pKa = 4.34DD58 pKa = 3.34EE59 pKa = 4.25RR60 pKa = 11.84RR61 pKa = 11.84GHH63 pKa = 5.94NMLRR67 pKa = 11.84EE68 pKa = 4.21FNSEE72 pKa = 3.91EE73 pKa = 4.37SGDD76 pKa = 3.82LPTFEE81 pKa = 6.61DD82 pKa = 3.71EE83 pKa = 5.43DD84 pKa = 4.6LFDD87 pKa = 4.58VEE89 pKa = 5.65LDD91 pKa = 3.76GEE93 pKa = 4.79GKK95 pKa = 8.1VTRR98 pKa = 11.84KK99 pKa = 9.55KK100 pKa = 9.95IRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84MHH106 pKa = 5.87KK107 pKa = 9.82PFVHH111 pKa = 6.22RR112 pKa = 11.84LVRR115 pKa = 11.84HH116 pKa = 5.65CRR118 pKa = 11.84GEE120 pKa = 3.81LGQRR124 pKa = 11.84VHH126 pKa = 6.57TPPNVMVVEE135 pKa = 4.85RR136 pKa = 11.84TARR139 pKa = 11.84AYY141 pKa = 10.46CHH143 pKa = 5.13EE144 pKa = 4.56HH145 pKa = 5.52YY146 pKa = 10.75VRR148 pKa = 11.84SSDD151 pKa = 3.2ISCILPVVVALYY163 pKa = 9.29FFSRR167 pKa = 11.84SDD169 pKa = 3.31VQIRR173 pKa = 11.84TEE175 pKa = 3.8ALKK178 pKa = 10.7QSSAFVKK185 pKa = 10.02SSKK188 pKa = 7.81PHH190 pKa = 6.0RR191 pKa = 11.84FVGVGGRR198 pKa = 11.84QHH200 pKa = 6.37GVADD204 pKa = 3.77AA205 pKa = 4.29
Molecular weight: 23.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
923 |
205 |
491 |
307.7 |
34.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.717 ± 0.818 | 1.95 ± 0.357 |
5.85 ± 0.717 | 4.875 ± 0.915 |
4.875 ± 0.346 | 6.501 ± 0.332 |
2.925 ± 0.71 | 5.309 ± 1.02 |
5.959 ± 0.759 | 7.259 ± 0.443 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.142 ± 0.422 | 4.984 ± 1.237 |
4.55 ± 0.257 | 2.6 ± 0.215 |
6.717 ± 1.593 | 8.126 ± 0.628 |
4.984 ± 1.469 | 7.692 ± 0.762 |
1.517 ± 0.252 | 3.467 ± 0.778 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
