
Salipiger aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Salipiger
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
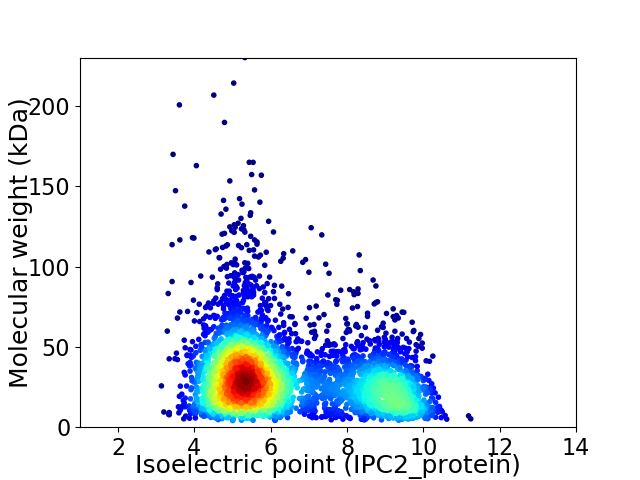
Virtual 2D-PAGE plot for 4452 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A327YEX1|A0A327YEX1_9RHOB Heterotetrameric sarcosine oxidase gamma subunit OS=Salipiger aestuarii OX=568098 GN=ATI53_1010103 PE=4 SV=1
MM1 pKa = 6.96ITRR4 pKa = 11.84FAIALAILGALSACNGNTSSGDD26 pKa = 3.71DD27 pKa = 3.48EE28 pKa = 4.81STDD31 pKa = 3.19EE32 pKa = 4.45DD33 pKa = 4.42TVEE36 pKa = 4.52EE37 pKa = 4.57PDD39 pKa = 3.97SFPLPGSRR47 pKa = 11.84VNPTASQSITRR58 pKa = 11.84MEE60 pKa = 4.16VDD62 pKa = 3.84DD63 pKa = 4.39GAGNGMIVDD72 pKa = 4.24PTYY75 pKa = 11.42DD76 pKa = 3.89PDD78 pKa = 3.79TDD80 pKa = 3.47TFYY83 pKa = 11.67VDD85 pKa = 4.28NLAFDD90 pKa = 4.38GANTYY95 pKa = 10.66ARR97 pKa = 11.84ADD99 pKa = 4.05FSPLAEE105 pKa = 4.05ITGALGPFAVYY116 pKa = 10.47EE117 pKa = 4.15NADD120 pKa = 3.79TVTDD124 pKa = 4.11PVNGDD129 pKa = 3.91VIPQLQHH136 pKa = 5.55KK137 pKa = 9.63AIYY140 pKa = 9.89AVSQSGQTEE149 pKa = 3.99LAIVRR154 pKa = 11.84TGAFVDD160 pKa = 3.7YY161 pKa = 10.88GFGGFVYY168 pKa = 10.2QRR170 pKa = 11.84NGGVTIPTSGQASYY184 pKa = 10.62SGSYY188 pKa = 10.32GGLRR192 pKa = 11.84DD193 pKa = 3.64YY194 pKa = 11.26QGTGKK199 pKa = 9.53IEE201 pKa = 4.23YY202 pKa = 8.47STADD206 pKa = 3.26AQVYY210 pKa = 10.38IDD212 pKa = 4.31FEE214 pKa = 5.05DD215 pKa = 4.16YY216 pKa = 11.5NEE218 pKa = 4.3GDD220 pKa = 3.86GVRR223 pKa = 11.84GAITDD228 pKa = 3.5RR229 pKa = 11.84RR230 pKa = 11.84IYY232 pKa = 10.64DD233 pKa = 3.52SNGVDD238 pKa = 3.27ISTDD242 pKa = 3.39YY243 pKa = 11.79VNIPTIQFEE252 pKa = 4.21IGPGVARR259 pKa = 11.84SNGEE263 pKa = 3.61MSGTAFSMVDD273 pKa = 3.28GATHH277 pKa = 5.5EE278 pKa = 4.45TGSFYY283 pKa = 11.36AVLSDD288 pKa = 4.32GADD291 pKa = 3.28NDD293 pKa = 3.67VTAEE297 pKa = 4.25EE298 pKa = 4.12IVGIIVIEE306 pKa = 4.24GTVGDD311 pKa = 3.91TLARR315 pKa = 11.84EE316 pKa = 4.28TGGFMVTRR324 pKa = 4.74
MM1 pKa = 6.96ITRR4 pKa = 11.84FAIALAILGALSACNGNTSSGDD26 pKa = 3.71DD27 pKa = 3.48EE28 pKa = 4.81STDD31 pKa = 3.19EE32 pKa = 4.45DD33 pKa = 4.42TVEE36 pKa = 4.52EE37 pKa = 4.57PDD39 pKa = 3.97SFPLPGSRR47 pKa = 11.84VNPTASQSITRR58 pKa = 11.84MEE60 pKa = 4.16VDD62 pKa = 3.84DD63 pKa = 4.39GAGNGMIVDD72 pKa = 4.24PTYY75 pKa = 11.42DD76 pKa = 3.89PDD78 pKa = 3.79TDD80 pKa = 3.47TFYY83 pKa = 11.67VDD85 pKa = 4.28NLAFDD90 pKa = 4.38GANTYY95 pKa = 10.66ARR97 pKa = 11.84ADD99 pKa = 4.05FSPLAEE105 pKa = 4.05ITGALGPFAVYY116 pKa = 10.47EE117 pKa = 4.15NADD120 pKa = 3.79TVTDD124 pKa = 4.11PVNGDD129 pKa = 3.91VIPQLQHH136 pKa = 5.55KK137 pKa = 9.63AIYY140 pKa = 9.89AVSQSGQTEE149 pKa = 3.99LAIVRR154 pKa = 11.84TGAFVDD160 pKa = 3.7YY161 pKa = 10.88GFGGFVYY168 pKa = 10.2QRR170 pKa = 11.84NGGVTIPTSGQASYY184 pKa = 10.62SGSYY188 pKa = 10.32GGLRR192 pKa = 11.84DD193 pKa = 3.64YY194 pKa = 11.26QGTGKK199 pKa = 9.53IEE201 pKa = 4.23YY202 pKa = 8.47STADD206 pKa = 3.26AQVYY210 pKa = 10.38IDD212 pKa = 4.31FEE214 pKa = 5.05DD215 pKa = 4.16YY216 pKa = 11.5NEE218 pKa = 4.3GDD220 pKa = 3.86GVRR223 pKa = 11.84GAITDD228 pKa = 3.5RR229 pKa = 11.84RR230 pKa = 11.84IYY232 pKa = 10.64DD233 pKa = 3.52SNGVDD238 pKa = 3.27ISTDD242 pKa = 3.39YY243 pKa = 11.79VNIPTIQFEE252 pKa = 4.21IGPGVARR259 pKa = 11.84SNGEE263 pKa = 3.61MSGTAFSMVDD273 pKa = 3.28GATHH277 pKa = 5.5EE278 pKa = 4.45TGSFYY283 pKa = 11.36AVLSDD288 pKa = 4.32GADD291 pKa = 3.28NDD293 pKa = 3.67VTAEE297 pKa = 4.25EE298 pKa = 4.12IVGIIVIEE306 pKa = 4.24GTVGDD311 pKa = 3.91TLARR315 pKa = 11.84EE316 pKa = 4.28TGGFMVTRR324 pKa = 4.74
Molecular weight: 34.16 kDa
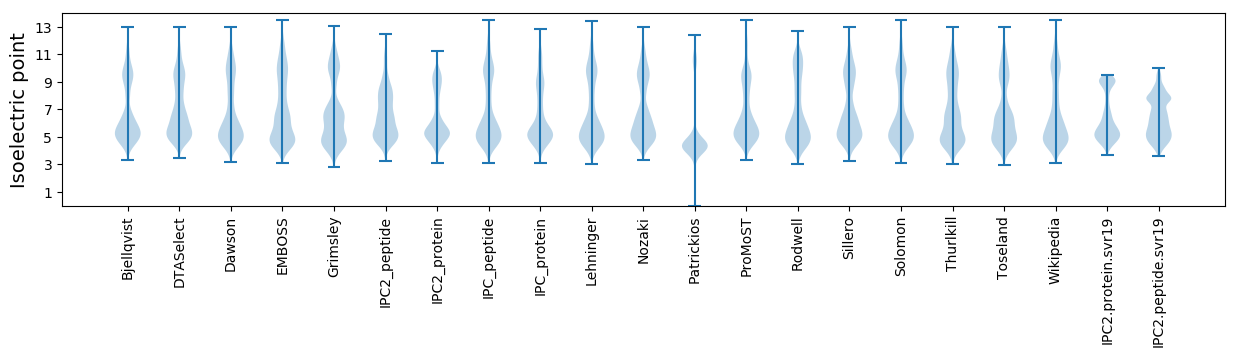
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A327XUZ6|A0A327XUZ6_9RHOB Tol-Pal system protein TolB OS=Salipiger aestuarii OX=568098 GN=tolB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.41ILNARR34 pKa = 11.84RR35 pKa = 11.84VRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.6QLSAA44 pKa = 3.97
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.41ILNARR34 pKa = 11.84RR35 pKa = 11.84VRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.6QLSAA44 pKa = 3.97
Molecular weight: 5.21 kDa
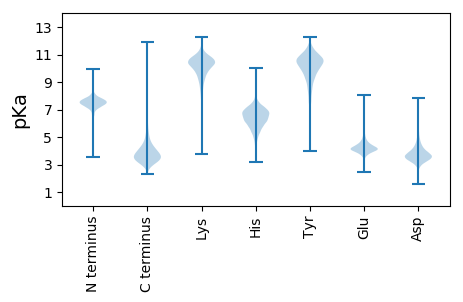
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1363853 |
39 |
2139 |
306.3 |
33.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.701 ± 0.057 | 0.948 ± 0.013 |
6.356 ± 0.038 | 5.324 ± 0.035 |
3.567 ± 0.025 | 8.907 ± 0.037 |
2.092 ± 0.02 | 4.888 ± 0.026 |
2.734 ± 0.029 | 10.106 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.777 ± 0.019 | 2.383 ± 0.021 |
5.298 ± 0.03 | 3.123 ± 0.019 |
7.334 ± 0.041 | 5.126 ± 0.027 |
5.536 ± 0.027 | 7.233 ± 0.035 |
1.428 ± 0.016 | 2.14 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
