
Riverside virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
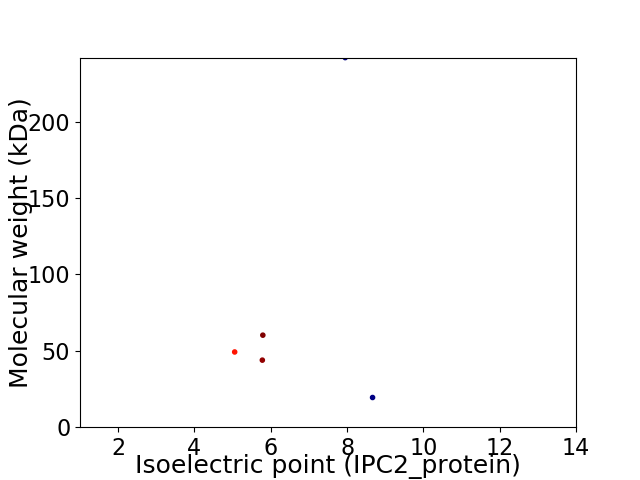
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140D049|A0A140D049_9RHAB Glycoprotein OS=Riverside virus 1 OX=1803263 GN=G PE=4 SV=1
MM1 pKa = 7.51AVNRR5 pKa = 11.84PIKK8 pKa = 10.14RR9 pKa = 11.84ISDD12 pKa = 3.7HH13 pKa = 7.19PEE15 pKa = 3.52LSPEE19 pKa = 4.14VVHH22 pKa = 7.09FDD24 pKa = 3.01IGTDD28 pKa = 3.43RR29 pKa = 11.84PIGYY33 pKa = 8.68PSAWFAQNVNDD44 pKa = 4.7KK45 pKa = 8.82PTIMMIPDD53 pKa = 3.75ASVTRR58 pKa = 11.84EE59 pKa = 3.86QLHH62 pKa = 5.2AAIRR66 pKa = 11.84AGLLTGTVDD75 pKa = 2.74HH76 pKa = 6.71RR77 pKa = 11.84FVVRR81 pKa = 11.84FLYY84 pKa = 10.99NEE86 pKa = 4.04FNLVPGLLNSDD97 pKa = 2.69WRR99 pKa = 11.84SFGRR103 pKa = 11.84NLGNRR108 pKa = 11.84GEE110 pKa = 4.54RR111 pKa = 11.84IGPSSIIVPRR121 pKa = 11.84EE122 pKa = 3.92QEE124 pKa = 4.24SVPALVAGNGLMDD137 pKa = 4.22DD138 pKa = 4.12QEE140 pKa = 4.33VLRR143 pKa = 11.84CIILICSVYY152 pKa = 10.07RR153 pKa = 11.84AGPISRR159 pKa = 11.84VDD161 pKa = 3.3YY162 pKa = 10.52RR163 pKa = 11.84DD164 pKa = 3.28QVLEE168 pKa = 4.04NVGNLLTPLGMDD180 pKa = 3.06QDD182 pKa = 4.01TDD184 pKa = 3.17IGDD187 pKa = 4.55IIDD190 pKa = 4.64KK191 pKa = 9.77CKK193 pKa = 10.43QWIAYY198 pKa = 8.08QPYY201 pKa = 7.75VQMMATIDD209 pKa = 3.44MFLNEE214 pKa = 4.98FPFHH218 pKa = 7.29PYY220 pKa = 8.59SQARR224 pKa = 11.84IGTIVTRR231 pKa = 11.84FKK233 pKa = 11.24DD234 pKa = 3.84CAALIATRR242 pKa = 11.84MITQCLGLKK251 pKa = 10.12FPEE254 pKa = 3.98FAEE257 pKa = 4.74WIWTSRR263 pKa = 11.84CADD266 pKa = 3.23QFEE269 pKa = 4.76RR270 pKa = 11.84IIKK273 pKa = 10.2GDD275 pKa = 3.55EE276 pKa = 4.06EE277 pKa = 3.97MDD279 pKa = 3.59EE280 pKa = 4.13PRR282 pKa = 11.84SYY284 pKa = 11.99SMYY287 pKa = 10.79FMDD290 pKa = 5.99LGLSSKK296 pKa = 10.5SPYY299 pKa = 9.2SASVNLDD306 pKa = 2.79LHH308 pKa = 6.51YY309 pKa = 10.58FFHH312 pKa = 6.68TLGVSAGLEE321 pKa = 3.74RR322 pKa = 11.84SKK324 pKa = 10.68RR325 pKa = 11.84ARR327 pKa = 11.84VVGDD331 pKa = 3.51PEE333 pKa = 4.28VNNIISNAVVMHH345 pKa = 5.47YY346 pKa = 11.37VMGRR350 pKa = 11.84FATLSQQFGVDD361 pKa = 3.57GEE363 pKa = 4.36GDD365 pKa = 3.56EE366 pKa = 6.52LEE368 pKa = 4.99DD369 pKa = 3.68EE370 pKa = 4.95NNRR373 pKa = 11.84PMPEE377 pKa = 4.24GEE379 pKa = 4.12PQEE382 pKa = 4.54KK383 pKa = 10.47DD384 pKa = 3.09SSLWLAYY391 pKa = 9.73IMRR394 pKa = 11.84NNGKK398 pKa = 9.25VPDD401 pKa = 4.08YY402 pKa = 10.51IVRR405 pKa = 11.84KK406 pKa = 9.65VATEE410 pKa = 4.23WEE412 pKa = 4.26QNPACRR418 pKa = 11.84DD419 pKa = 3.55GTIGKK424 pKa = 9.39ALYY427 pKa = 10.14DD428 pKa = 3.36KK429 pKa = 11.11SGPLLLNN436 pKa = 4.1
MM1 pKa = 7.51AVNRR5 pKa = 11.84PIKK8 pKa = 10.14RR9 pKa = 11.84ISDD12 pKa = 3.7HH13 pKa = 7.19PEE15 pKa = 3.52LSPEE19 pKa = 4.14VVHH22 pKa = 7.09FDD24 pKa = 3.01IGTDD28 pKa = 3.43RR29 pKa = 11.84PIGYY33 pKa = 8.68PSAWFAQNVNDD44 pKa = 4.7KK45 pKa = 8.82PTIMMIPDD53 pKa = 3.75ASVTRR58 pKa = 11.84EE59 pKa = 3.86QLHH62 pKa = 5.2AAIRR66 pKa = 11.84AGLLTGTVDD75 pKa = 2.74HH76 pKa = 6.71RR77 pKa = 11.84FVVRR81 pKa = 11.84FLYY84 pKa = 10.99NEE86 pKa = 4.04FNLVPGLLNSDD97 pKa = 2.69WRR99 pKa = 11.84SFGRR103 pKa = 11.84NLGNRR108 pKa = 11.84GEE110 pKa = 4.54RR111 pKa = 11.84IGPSSIIVPRR121 pKa = 11.84EE122 pKa = 3.92QEE124 pKa = 4.24SVPALVAGNGLMDD137 pKa = 4.22DD138 pKa = 4.12QEE140 pKa = 4.33VLRR143 pKa = 11.84CIILICSVYY152 pKa = 10.07RR153 pKa = 11.84AGPISRR159 pKa = 11.84VDD161 pKa = 3.3YY162 pKa = 10.52RR163 pKa = 11.84DD164 pKa = 3.28QVLEE168 pKa = 4.04NVGNLLTPLGMDD180 pKa = 3.06QDD182 pKa = 4.01TDD184 pKa = 3.17IGDD187 pKa = 4.55IIDD190 pKa = 4.64KK191 pKa = 9.77CKK193 pKa = 10.43QWIAYY198 pKa = 8.08QPYY201 pKa = 7.75VQMMATIDD209 pKa = 3.44MFLNEE214 pKa = 4.98FPFHH218 pKa = 7.29PYY220 pKa = 8.59SQARR224 pKa = 11.84IGTIVTRR231 pKa = 11.84FKK233 pKa = 11.24DD234 pKa = 3.84CAALIATRR242 pKa = 11.84MITQCLGLKK251 pKa = 10.12FPEE254 pKa = 3.98FAEE257 pKa = 4.74WIWTSRR263 pKa = 11.84CADD266 pKa = 3.23QFEE269 pKa = 4.76RR270 pKa = 11.84IIKK273 pKa = 10.2GDD275 pKa = 3.55EE276 pKa = 4.06EE277 pKa = 3.97MDD279 pKa = 3.59EE280 pKa = 4.13PRR282 pKa = 11.84SYY284 pKa = 11.99SMYY287 pKa = 10.79FMDD290 pKa = 5.99LGLSSKK296 pKa = 10.5SPYY299 pKa = 9.2SASVNLDD306 pKa = 2.79LHH308 pKa = 6.51YY309 pKa = 10.58FFHH312 pKa = 6.68TLGVSAGLEE321 pKa = 3.74RR322 pKa = 11.84SKK324 pKa = 10.68RR325 pKa = 11.84ARR327 pKa = 11.84VVGDD331 pKa = 3.51PEE333 pKa = 4.28VNNIISNAVVMHH345 pKa = 5.47YY346 pKa = 11.37VMGRR350 pKa = 11.84FATLSQQFGVDD361 pKa = 3.57GEE363 pKa = 4.36GDD365 pKa = 3.56EE366 pKa = 6.52LEE368 pKa = 4.99DD369 pKa = 3.68EE370 pKa = 4.95NNRR373 pKa = 11.84PMPEE377 pKa = 4.24GEE379 pKa = 4.12PQEE382 pKa = 4.54KK383 pKa = 10.47DD384 pKa = 3.09SSLWLAYY391 pKa = 9.73IMRR394 pKa = 11.84NNGKK398 pKa = 9.25VPDD401 pKa = 4.08YY402 pKa = 10.51IVRR405 pKa = 11.84KK406 pKa = 9.65VATEE410 pKa = 4.23WEE412 pKa = 4.26QNPACRR418 pKa = 11.84DD419 pKa = 3.55GTIGKK424 pKa = 9.39ALYY427 pKa = 10.14DD428 pKa = 3.36KK429 pKa = 11.11SGPLLLNN436 pKa = 4.1
Molecular weight: 49.26 kDa
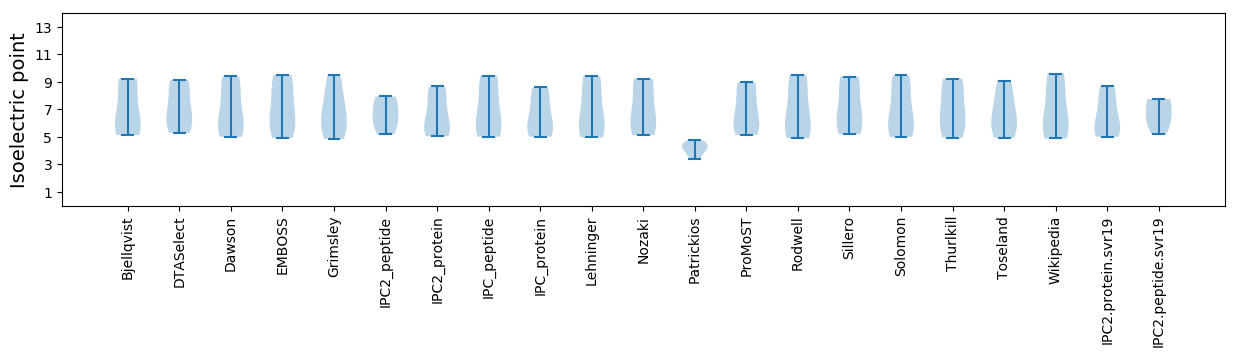
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140D058|A0A140D058_9RHAB Matrix protein OS=Riverside virus 1 OX=1803263 GN=M PE=4 SV=1
MM1 pKa = 7.54SIIKK5 pKa = 9.9IVRR8 pKa = 11.84VDD10 pKa = 3.5ALLEE14 pKa = 3.86MRR16 pKa = 11.84FRR18 pKa = 11.84EE19 pKa = 4.22PPRR22 pKa = 11.84TYY24 pKa = 11.07KK25 pKa = 10.74DD26 pKa = 3.47LLSSLEE32 pKa = 4.24IIKK35 pKa = 10.43DD36 pKa = 3.57DD37 pKa = 3.56YY38 pKa = 11.23KK39 pKa = 11.52GYY41 pKa = 11.24VNDD44 pKa = 4.41KK45 pKa = 10.59EE46 pKa = 4.47YY47 pKa = 10.8ILLFYY52 pKa = 9.94TLAGFTCKK60 pKa = 9.9YY61 pKa = 8.1RR62 pKa = 11.84HH63 pKa = 5.2QVGAFYY69 pKa = 10.26HH70 pKa = 5.42YY71 pKa = 10.13RR72 pKa = 11.84GTINGVFKK80 pKa = 10.75IKK82 pKa = 9.11MCSMSPIFPLTYY94 pKa = 10.09DD95 pKa = 3.61YY96 pKa = 11.4DD97 pKa = 4.28GEE99 pKa = 4.86GIGLIPNLLVSFHH112 pKa = 6.12LTSEE116 pKa = 4.28EE117 pKa = 4.7SNMSPTSIVDD127 pKa = 3.72YY128 pKa = 10.64IRR130 pKa = 11.84SRR132 pKa = 11.84SPNKK136 pKa = 9.09YY137 pKa = 9.55RR138 pKa = 11.84KK139 pKa = 9.02RR140 pKa = 11.84HH141 pKa = 4.89ALKK144 pKa = 10.74LSIFKK149 pKa = 10.19PYY151 pKa = 10.46AITVEE156 pKa = 4.19SEE158 pKa = 3.95KK159 pKa = 11.18DD160 pKa = 3.3CLSIKK165 pKa = 10.65KK166 pKa = 9.84NN167 pKa = 3.41
MM1 pKa = 7.54SIIKK5 pKa = 9.9IVRR8 pKa = 11.84VDD10 pKa = 3.5ALLEE14 pKa = 3.86MRR16 pKa = 11.84FRR18 pKa = 11.84EE19 pKa = 4.22PPRR22 pKa = 11.84TYY24 pKa = 11.07KK25 pKa = 10.74DD26 pKa = 3.47LLSSLEE32 pKa = 4.24IIKK35 pKa = 10.43DD36 pKa = 3.57DD37 pKa = 3.56YY38 pKa = 11.23KK39 pKa = 11.52GYY41 pKa = 11.24VNDD44 pKa = 4.41KK45 pKa = 10.59EE46 pKa = 4.47YY47 pKa = 10.8ILLFYY52 pKa = 9.94TLAGFTCKK60 pKa = 9.9YY61 pKa = 8.1RR62 pKa = 11.84HH63 pKa = 5.2QVGAFYY69 pKa = 10.26HH70 pKa = 5.42YY71 pKa = 10.13RR72 pKa = 11.84GTINGVFKK80 pKa = 10.75IKK82 pKa = 9.11MCSMSPIFPLTYY94 pKa = 10.09DD95 pKa = 3.61YY96 pKa = 11.4DD97 pKa = 4.28GEE99 pKa = 4.86GIGLIPNLLVSFHH112 pKa = 6.12LTSEE116 pKa = 4.28EE117 pKa = 4.7SNMSPTSIVDD127 pKa = 3.72YY128 pKa = 10.64IRR130 pKa = 11.84SRR132 pKa = 11.84SPNKK136 pKa = 9.09YY137 pKa = 9.55RR138 pKa = 11.84KK139 pKa = 9.02RR140 pKa = 11.84HH141 pKa = 4.89ALKK144 pKa = 10.74LSIFKK149 pKa = 10.19PYY151 pKa = 10.46AITVEE156 pKa = 4.19SEE158 pKa = 3.95KK159 pKa = 11.18DD160 pKa = 3.3CLSIKK165 pKa = 10.65KK166 pKa = 9.84NN167 pKa = 3.41
Molecular weight: 19.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3656 |
167 |
2122 |
731.2 |
82.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.787 ± 0.74 | 1.422 ± 0.278 |
5.771 ± 0.356 | 5.388 ± 0.714 |
4.212 ± 0.173 | 6.072 ± 0.459 |
2.571 ± 0.436 | 6.838 ± 0.544 |
6.154 ± 0.637 | 9.874 ± 0.863 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.34 | 4.814 ± 0.353 |
5.17 ± 0.62 | 3.556 ± 0.44 |
5.142 ± 0.467 | 8.124 ± 0.755 |
6.209 ± 0.679 | 5.826 ± 0.364 |
1.723 ± 0.393 | 3.857 ± 0.673 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
