
endosymbiont of Tevnia jerichonana (vent Tica)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; sulfur-oxidizing symbionts; endosymbiont of Tevnia jerichonana
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
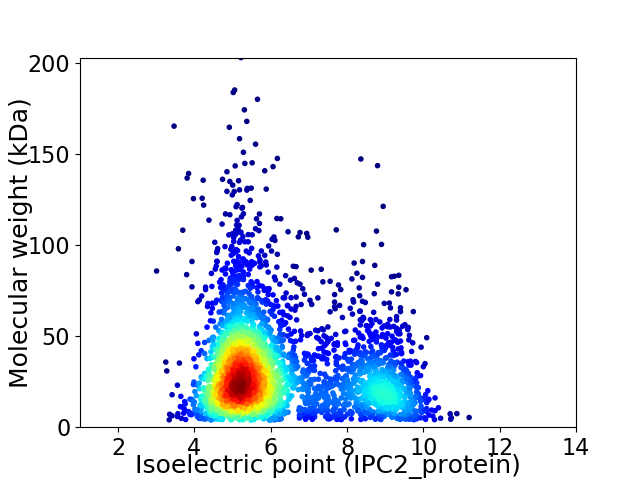
Virtual 2D-PAGE plot for 3230 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
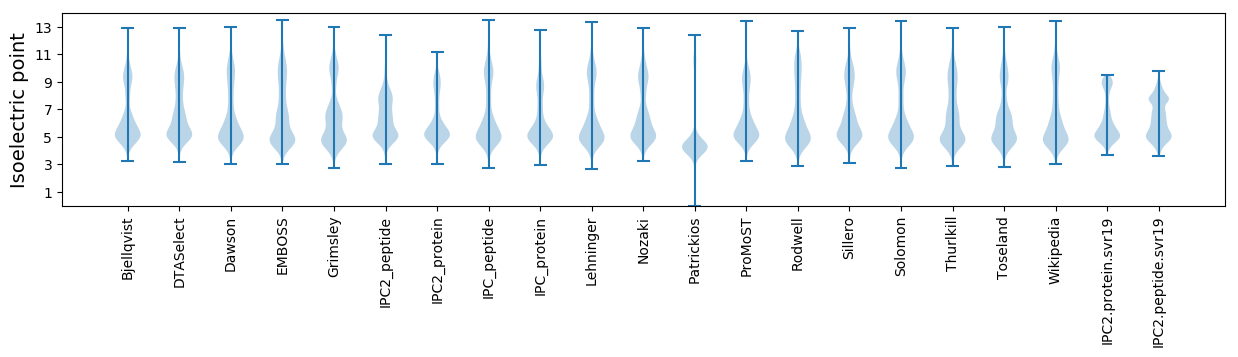
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2FJL0|G2FJL0_9GAMM Electron transport complex protein RnfA OS=endosymbiont of Tevnia jerichonana (vent Tica) OX=1049564 GN=rnfA3 PE=4 SV=1
MM1 pKa = 7.51LFAGFSVFAEE11 pKa = 4.08VLVEE15 pKa = 5.48DD16 pKa = 5.37GFDD19 pKa = 3.38NDD21 pKa = 3.99SAGFTYY27 pKa = 10.71VDD29 pKa = 4.25DD30 pKa = 4.9PFHH33 pKa = 6.81GSSAPGYY40 pKa = 10.72ADD42 pKa = 3.72GSHH45 pKa = 6.74LSVGGYY51 pKa = 9.62SGGALQVILGGRR63 pKa = 11.84DD64 pKa = 3.3TIDD67 pKa = 3.41LFGLSGGWQQEE78 pKa = 4.1FEE80 pKa = 4.29LAGLSEE86 pKa = 4.33VSLSFRR92 pKa = 11.84YY93 pKa = 9.83KK94 pKa = 10.35LSQSPFYY101 pKa = 10.91EE102 pKa = 4.63GDD104 pKa = 3.35EE105 pKa = 4.17YY106 pKa = 11.68SQVLVSIDD114 pKa = 3.36GVLVGVSPNDD124 pKa = 3.74YY125 pKa = 10.01IDD127 pKa = 3.58QVSGNGNGGAEE138 pKa = 4.29EE139 pKa = 4.35TTGWRR144 pKa = 11.84QVEE147 pKa = 4.34LNLGGLAAGRR157 pKa = 11.84HH158 pKa = 4.72RR159 pKa = 11.84LIIGGYY165 pKa = 9.2NSQKK169 pKa = 8.91TAANEE174 pKa = 4.13TTEE177 pKa = 4.27VLIDD181 pKa = 4.35DD182 pKa = 5.31LLLSSGNSNLPPRR195 pKa = 11.84TVVGADD201 pKa = 3.33PTAGVAPLQVQFSSEE216 pKa = 3.92GSTDD220 pKa = 3.3PDD222 pKa = 3.35GRR224 pKa = 11.84IVGYY228 pKa = 9.74HH229 pKa = 4.86WVFGDD234 pKa = 3.97GNSSTEE240 pKa = 3.69INPLHH245 pKa = 6.88SYY247 pKa = 8.15ATAGNYY253 pKa = 7.39TVTLILTDD261 pKa = 3.73DD262 pKa = 3.49QGATGSASIMINVSGLSDD280 pKa = 3.45TAAPEE285 pKa = 3.86IPAGVSAIPSGADD298 pKa = 3.21TIQLSWQAASDD309 pKa = 3.95NVGVTGYY316 pKa = 10.65RR317 pKa = 11.84VFRR320 pKa = 11.84DD321 pKa = 3.76GVEE324 pKa = 4.11IASSTTTSYY333 pKa = 11.45RR334 pKa = 11.84DD335 pKa = 3.33TGLSPLTRR343 pKa = 11.84YY344 pKa = 9.61SYY346 pKa = 10.73RR347 pKa = 11.84VSAYY351 pKa = 9.96DD352 pKa = 3.24AAGNEE357 pKa = 4.33SEE359 pKa = 4.25PGEE362 pKa = 4.28VVAATTGNQLVPLIEE377 pKa = 4.65SGFDD381 pKa = 3.17SDD383 pKa = 4.42SAGFSYY389 pKa = 11.3VDD391 pKa = 3.37DD392 pKa = 4.41PFRR395 pKa = 11.84NTSAPGYY402 pKa = 10.85ADD404 pKa = 3.32GSYY407 pKa = 10.35LGAGGYY413 pKa = 9.56SGSALQITLGGIDD426 pKa = 3.93GDD428 pKa = 5.09DD429 pKa = 3.21IFGLSGGWQRR439 pKa = 11.84DD440 pKa = 3.85FEE442 pKa = 4.46LTSQSEE448 pKa = 4.19LSLSFRR454 pKa = 11.84YY455 pKa = 9.7RR456 pKa = 11.84LSQSPFYY463 pKa = 10.57EE464 pKa = 3.74QDD466 pKa = 3.24EE467 pKa = 4.3QSQLLVSIDD476 pKa = 3.46GVLVGVSPNDD486 pKa = 3.68YY487 pKa = 10.15IDD489 pKa = 4.02QISGNGNGGAEE500 pKa = 4.29EE501 pKa = 4.35TTGWRR506 pKa = 11.84QVALSLGSLVGGSHH520 pKa = 6.81RR521 pKa = 11.84LIIGGYY527 pKa = 9.08NNQKK531 pKa = 7.95TAANEE536 pKa = 4.14TTEE539 pKa = 4.13VLIDD543 pKa = 3.98DD544 pKa = 4.52LVLRR548 pKa = 11.84SDD550 pKa = 4.2SSNLPPSAVASAAPTAGIAPLEE572 pKa = 4.18VSFSSVGSSDD582 pKa = 3.58PDD584 pKa = 3.23GTIQHH589 pKa = 6.49FSWAFGDD596 pKa = 4.44GSSSTEE602 pKa = 3.6ANPLHH607 pKa = 7.14SYY609 pKa = 6.96TTPGDD614 pKa = 3.58YY615 pKa = 9.39TATLSVTDD623 pKa = 3.89NQGATASASVGISVSDD639 pKa = 4.29LPDD642 pKa = 3.45TEE644 pKa = 4.96APEE647 pKa = 4.31IPAGLSAIPSGSDD660 pKa = 3.18TIEE663 pKa = 4.64LSWQAASDD671 pKa = 3.95NVGVTGYY678 pKa = 10.65RR679 pKa = 11.84VFRR682 pKa = 11.84DD683 pKa = 3.76GVEE686 pKa = 4.0IASSATISYY695 pKa = 10.0RR696 pKa = 11.84DD697 pKa = 3.45SRR699 pKa = 11.84LTPSTRR705 pKa = 11.84YY706 pKa = 9.61SYY708 pKa = 10.79QISAYY713 pKa = 9.93DD714 pKa = 3.27AAGNQSEE721 pKa = 4.58PGVAVAATTGAQPTLTSLSLTPASAQLNIGLRR753 pKa = 11.84QQFNVSGFDD762 pKa = 3.28QYY764 pKa = 12.17GNTIVVNPTWSVNGGGVIDD783 pKa = 3.77STGLFSASQAGGPFTVLAEE802 pKa = 4.12QDD804 pKa = 3.7GVRR807 pKa = 11.84ATASLTVIRR816 pKa = 11.84QSNLIDD822 pKa = 3.54SGFDD826 pKa = 3.13SDD828 pKa = 4.76SAGFSYY834 pKa = 11.09LDD836 pKa = 3.51DD837 pKa = 4.37PFRR840 pKa = 11.84GSGAPAYY847 pKa = 10.5ADD849 pKa = 3.3GSYY852 pKa = 10.45LGAGGYY858 pKa = 9.84SGGALQITLGGIDD871 pKa = 3.95GDD873 pKa = 5.25DD874 pKa = 3.33IFGMSGGWQHH884 pKa = 7.26DD885 pKa = 3.77FQLNALSEE893 pKa = 4.35VSLSFRR899 pKa = 11.84YY900 pKa = 9.71QLSQTPFYY908 pKa = 10.54EE909 pKa = 4.15RR910 pKa = 11.84NEE912 pKa = 3.88YY913 pKa = 10.33SQLLLSIDD921 pKa = 3.62GVLVGVSPNDD931 pKa = 4.07FIDD934 pKa = 3.96QISGNGNGGAEE945 pKa = 4.29EE946 pKa = 4.35TTGWRR951 pKa = 11.84QVEE954 pKa = 4.35LNLGSLAAGSHH965 pKa = 5.78RR966 pKa = 11.84LIIGAYY972 pKa = 9.29NNQKK976 pKa = 7.97TAANEE981 pKa = 4.23TTVVLIDD988 pKa = 4.24DD989 pKa = 4.57LLLRR993 pKa = 11.84SSSSLPVNLPPSAVASADD1011 pKa = 3.47PQTGVVPLVVNFSSAGSTDD1030 pKa = 3.42PDD1032 pKa = 3.48GTIEE1036 pKa = 4.1HH1037 pKa = 6.31VSWAFGDD1044 pKa = 4.45GSSSTEE1050 pKa = 3.6ANPLHH1055 pKa = 7.14SYY1057 pKa = 6.96TTPGDD1062 pKa = 3.58YY1063 pKa = 9.39TATLSVTDD1071 pKa = 3.89NQGATASASVGISVSDD1087 pKa = 3.9QPDD1090 pKa = 3.53TEE1092 pKa = 4.37APEE1095 pKa = 4.27IPAGLSAIPSGSDD1108 pKa = 2.89IIEE1111 pKa = 4.62LSWQAASDD1119 pKa = 3.68NVGIIGYY1126 pKa = 9.4RR1127 pKa = 11.84VFRR1130 pKa = 11.84DD1131 pKa = 3.61GVEE1134 pKa = 3.97IANSAITSYY1143 pKa = 10.65RR1144 pKa = 11.84DD1145 pKa = 3.12SGLTPSTRR1153 pKa = 11.84YY1154 pKa = 9.67SYY1156 pKa = 10.74QVSAYY1161 pKa = 9.92DD1162 pKa = 3.33AAGNQSEE1169 pKa = 4.58PGVAVAATTGNQPSLLIEE1187 pKa = 5.19DD1188 pKa = 4.96GFDD1191 pKa = 3.2TDD1193 pKa = 3.85SAGFSYY1199 pKa = 11.22VDD1201 pKa = 3.38DD1202 pKa = 4.48PFRR1205 pKa = 11.84GSSAPGYY1212 pKa = 10.82ADD1214 pKa = 4.77GSLLPDD1220 pKa = 3.9GGYY1223 pKa = 10.36SGGALQIILGGINSADD1239 pKa = 3.03IFGLSGGWQRR1249 pKa = 11.84DD1250 pKa = 3.6FQLNGLSKK1258 pKa = 10.36VVLSFRR1264 pKa = 11.84YY1265 pKa = 9.56RR1266 pKa = 11.84LSQSPFYY1273 pKa = 10.57EE1274 pKa = 3.74QDD1276 pKa = 3.24EE1277 pKa = 4.3QSQLLVSLDD1286 pKa = 3.61GEE1288 pKa = 4.96LIGVTPNDD1296 pKa = 4.46YY1297 pKa = 10.45IDD1299 pKa = 3.59QVQGNGNGGPEE1310 pKa = 4.35EE1311 pKa = 4.32STGWRR1316 pKa = 11.84QVEE1319 pKa = 4.0LDD1321 pKa = 4.39LGNLAIGSHH1330 pKa = 5.71RR1331 pKa = 11.84QPSADD1336 pKa = 3.62HH1337 pKa = 6.99RR1338 pKa = 11.84RR1339 pKa = 11.84LQQQ1342 pKa = 3.09
MM1 pKa = 7.51LFAGFSVFAEE11 pKa = 4.08VLVEE15 pKa = 5.48DD16 pKa = 5.37GFDD19 pKa = 3.38NDD21 pKa = 3.99SAGFTYY27 pKa = 10.71VDD29 pKa = 4.25DD30 pKa = 4.9PFHH33 pKa = 6.81GSSAPGYY40 pKa = 10.72ADD42 pKa = 3.72GSHH45 pKa = 6.74LSVGGYY51 pKa = 9.62SGGALQVILGGRR63 pKa = 11.84DD64 pKa = 3.3TIDD67 pKa = 3.41LFGLSGGWQQEE78 pKa = 4.1FEE80 pKa = 4.29LAGLSEE86 pKa = 4.33VSLSFRR92 pKa = 11.84YY93 pKa = 9.83KK94 pKa = 10.35LSQSPFYY101 pKa = 10.91EE102 pKa = 4.63GDD104 pKa = 3.35EE105 pKa = 4.17YY106 pKa = 11.68SQVLVSIDD114 pKa = 3.36GVLVGVSPNDD124 pKa = 3.74YY125 pKa = 10.01IDD127 pKa = 3.58QVSGNGNGGAEE138 pKa = 4.29EE139 pKa = 4.35TTGWRR144 pKa = 11.84QVEE147 pKa = 4.34LNLGGLAAGRR157 pKa = 11.84HH158 pKa = 4.72RR159 pKa = 11.84LIIGGYY165 pKa = 9.2NSQKK169 pKa = 8.91TAANEE174 pKa = 4.13TTEE177 pKa = 4.27VLIDD181 pKa = 4.35DD182 pKa = 5.31LLLSSGNSNLPPRR195 pKa = 11.84TVVGADD201 pKa = 3.33PTAGVAPLQVQFSSEE216 pKa = 3.92GSTDD220 pKa = 3.3PDD222 pKa = 3.35GRR224 pKa = 11.84IVGYY228 pKa = 9.74HH229 pKa = 4.86WVFGDD234 pKa = 3.97GNSSTEE240 pKa = 3.69INPLHH245 pKa = 6.88SYY247 pKa = 8.15ATAGNYY253 pKa = 7.39TVTLILTDD261 pKa = 3.73DD262 pKa = 3.49QGATGSASIMINVSGLSDD280 pKa = 3.45TAAPEE285 pKa = 3.86IPAGVSAIPSGADD298 pKa = 3.21TIQLSWQAASDD309 pKa = 3.95NVGVTGYY316 pKa = 10.65RR317 pKa = 11.84VFRR320 pKa = 11.84DD321 pKa = 3.76GVEE324 pKa = 4.11IASSTTTSYY333 pKa = 11.45RR334 pKa = 11.84DD335 pKa = 3.33TGLSPLTRR343 pKa = 11.84YY344 pKa = 9.61SYY346 pKa = 10.73RR347 pKa = 11.84VSAYY351 pKa = 9.96DD352 pKa = 3.24AAGNEE357 pKa = 4.33SEE359 pKa = 4.25PGEE362 pKa = 4.28VVAATTGNQLVPLIEE377 pKa = 4.65SGFDD381 pKa = 3.17SDD383 pKa = 4.42SAGFSYY389 pKa = 11.3VDD391 pKa = 3.37DD392 pKa = 4.41PFRR395 pKa = 11.84NTSAPGYY402 pKa = 10.85ADD404 pKa = 3.32GSYY407 pKa = 10.35LGAGGYY413 pKa = 9.56SGSALQITLGGIDD426 pKa = 3.93GDD428 pKa = 5.09DD429 pKa = 3.21IFGLSGGWQRR439 pKa = 11.84DD440 pKa = 3.85FEE442 pKa = 4.46LTSQSEE448 pKa = 4.19LSLSFRR454 pKa = 11.84YY455 pKa = 9.7RR456 pKa = 11.84LSQSPFYY463 pKa = 10.57EE464 pKa = 3.74QDD466 pKa = 3.24EE467 pKa = 4.3QSQLLVSIDD476 pKa = 3.46GVLVGVSPNDD486 pKa = 3.68YY487 pKa = 10.15IDD489 pKa = 4.02QISGNGNGGAEE500 pKa = 4.29EE501 pKa = 4.35TTGWRR506 pKa = 11.84QVALSLGSLVGGSHH520 pKa = 6.81RR521 pKa = 11.84LIIGGYY527 pKa = 9.08NNQKK531 pKa = 7.95TAANEE536 pKa = 4.14TTEE539 pKa = 4.13VLIDD543 pKa = 3.98DD544 pKa = 4.52LVLRR548 pKa = 11.84SDD550 pKa = 4.2SSNLPPSAVASAAPTAGIAPLEE572 pKa = 4.18VSFSSVGSSDD582 pKa = 3.58PDD584 pKa = 3.23GTIQHH589 pKa = 6.49FSWAFGDD596 pKa = 4.44GSSSTEE602 pKa = 3.6ANPLHH607 pKa = 7.14SYY609 pKa = 6.96TTPGDD614 pKa = 3.58YY615 pKa = 9.39TATLSVTDD623 pKa = 3.89NQGATASASVGISVSDD639 pKa = 4.29LPDD642 pKa = 3.45TEE644 pKa = 4.96APEE647 pKa = 4.31IPAGLSAIPSGSDD660 pKa = 3.18TIEE663 pKa = 4.64LSWQAASDD671 pKa = 3.95NVGVTGYY678 pKa = 10.65RR679 pKa = 11.84VFRR682 pKa = 11.84DD683 pKa = 3.76GVEE686 pKa = 4.0IASSATISYY695 pKa = 10.0RR696 pKa = 11.84DD697 pKa = 3.45SRR699 pKa = 11.84LTPSTRR705 pKa = 11.84YY706 pKa = 9.61SYY708 pKa = 10.79QISAYY713 pKa = 9.93DD714 pKa = 3.27AAGNQSEE721 pKa = 4.58PGVAVAATTGAQPTLTSLSLTPASAQLNIGLRR753 pKa = 11.84QQFNVSGFDD762 pKa = 3.28QYY764 pKa = 12.17GNTIVVNPTWSVNGGGVIDD783 pKa = 3.77STGLFSASQAGGPFTVLAEE802 pKa = 4.12QDD804 pKa = 3.7GVRR807 pKa = 11.84ATASLTVIRR816 pKa = 11.84QSNLIDD822 pKa = 3.54SGFDD826 pKa = 3.13SDD828 pKa = 4.76SAGFSYY834 pKa = 11.09LDD836 pKa = 3.51DD837 pKa = 4.37PFRR840 pKa = 11.84GSGAPAYY847 pKa = 10.5ADD849 pKa = 3.3GSYY852 pKa = 10.45LGAGGYY858 pKa = 9.84SGGALQITLGGIDD871 pKa = 3.95GDD873 pKa = 5.25DD874 pKa = 3.33IFGMSGGWQHH884 pKa = 7.26DD885 pKa = 3.77FQLNALSEE893 pKa = 4.35VSLSFRR899 pKa = 11.84YY900 pKa = 9.71QLSQTPFYY908 pKa = 10.54EE909 pKa = 4.15RR910 pKa = 11.84NEE912 pKa = 3.88YY913 pKa = 10.33SQLLLSIDD921 pKa = 3.62GVLVGVSPNDD931 pKa = 4.07FIDD934 pKa = 3.96QISGNGNGGAEE945 pKa = 4.29EE946 pKa = 4.35TTGWRR951 pKa = 11.84QVEE954 pKa = 4.35LNLGSLAAGSHH965 pKa = 5.78RR966 pKa = 11.84LIIGAYY972 pKa = 9.29NNQKK976 pKa = 7.97TAANEE981 pKa = 4.23TTVVLIDD988 pKa = 4.24DD989 pKa = 4.57LLLRR993 pKa = 11.84SSSSLPVNLPPSAVASADD1011 pKa = 3.47PQTGVVPLVVNFSSAGSTDD1030 pKa = 3.42PDD1032 pKa = 3.48GTIEE1036 pKa = 4.1HH1037 pKa = 6.31VSWAFGDD1044 pKa = 4.45GSSSTEE1050 pKa = 3.6ANPLHH1055 pKa = 7.14SYY1057 pKa = 6.96TTPGDD1062 pKa = 3.58YY1063 pKa = 9.39TATLSVTDD1071 pKa = 3.89NQGATASASVGISVSDD1087 pKa = 3.9QPDD1090 pKa = 3.53TEE1092 pKa = 4.37APEE1095 pKa = 4.27IPAGLSAIPSGSDD1108 pKa = 2.89IIEE1111 pKa = 4.62LSWQAASDD1119 pKa = 3.68NVGIIGYY1126 pKa = 9.4RR1127 pKa = 11.84VFRR1130 pKa = 11.84DD1131 pKa = 3.61GVEE1134 pKa = 3.97IANSAITSYY1143 pKa = 10.65RR1144 pKa = 11.84DD1145 pKa = 3.12SGLTPSTRR1153 pKa = 11.84YY1154 pKa = 9.67SYY1156 pKa = 10.74QVSAYY1161 pKa = 9.92DD1162 pKa = 3.33AAGNQSEE1169 pKa = 4.58PGVAVAATTGNQPSLLIEE1187 pKa = 5.19DD1188 pKa = 4.96GFDD1191 pKa = 3.2TDD1193 pKa = 3.85SAGFSYY1199 pKa = 11.22VDD1201 pKa = 3.38DD1202 pKa = 4.48PFRR1205 pKa = 11.84GSSAPGYY1212 pKa = 10.82ADD1214 pKa = 4.77GSLLPDD1220 pKa = 3.9GGYY1223 pKa = 10.36SGGALQIILGGINSADD1239 pKa = 3.03IFGLSGGWQRR1249 pKa = 11.84DD1250 pKa = 3.6FQLNGLSKK1258 pKa = 10.36VVLSFRR1264 pKa = 11.84YY1265 pKa = 9.56RR1266 pKa = 11.84LSQSPFYY1273 pKa = 10.57EE1274 pKa = 3.74QDD1276 pKa = 3.24EE1277 pKa = 4.3QSQLLVSLDD1286 pKa = 3.61GEE1288 pKa = 4.96LIGVTPNDD1296 pKa = 4.46YY1297 pKa = 10.45IDD1299 pKa = 3.59QVQGNGNGGPEE1310 pKa = 4.35EE1311 pKa = 4.32STGWRR1316 pKa = 11.84QVEE1319 pKa = 4.0LDD1321 pKa = 4.39LGNLAIGSHH1330 pKa = 5.71RR1331 pKa = 11.84QPSADD1336 pKa = 3.62HH1337 pKa = 6.99RR1338 pKa = 11.84RR1339 pKa = 11.84LQQQ1342 pKa = 3.09
Molecular weight: 139.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2FGQ4|G2FGQ4_9GAMM Kinesin light chain-like protein OS=endosymbiont of Tevnia jerichonana (vent Tica) OX=1049564 GN=TevJSym_ar00330 PE=4 SV=1
MM1 pKa = 7.45MKK3 pKa = 9.48RR4 pKa = 11.84TFQPSNLKK12 pKa = 9.99RR13 pKa = 11.84VRR15 pKa = 11.84THH17 pKa = 6.07GFRR20 pKa = 11.84TRR22 pKa = 11.84MATQNGRR29 pKa = 11.84KK30 pKa = 9.09VIKK33 pKa = 10.14ARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.69GRR40 pKa = 11.84ARR42 pKa = 11.84LIPP45 pKa = 4.0
MM1 pKa = 7.45MKK3 pKa = 9.48RR4 pKa = 11.84TFQPSNLKK12 pKa = 9.99RR13 pKa = 11.84VRR15 pKa = 11.84THH17 pKa = 6.07GFRR20 pKa = 11.84TRR22 pKa = 11.84MATQNGRR29 pKa = 11.84KK30 pKa = 9.09VIKK33 pKa = 10.14ARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.69GRR40 pKa = 11.84ARR42 pKa = 11.84LIPP45 pKa = 4.0
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1002472 |
37 |
1870 |
310.4 |
34.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.749 ± 0.046 | 1.004 ± 0.02 |
5.507 ± 0.036 | 6.725 ± 0.044 |
3.607 ± 0.027 | 7.768 ± 0.041 |
2.323 ± 0.025 | 5.55 ± 0.035 |
3.677 ± 0.042 | 11.38 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.499 ± 0.021 | 3.101 ± 0.031 |
4.706 ± 0.033 | 4.734 ± 0.044 |
6.655 ± 0.044 | 6.054 ± 0.041 |
4.621 ± 0.036 | 6.535 ± 0.043 |
1.283 ± 0.018 | 2.523 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
