
Human adenovirus C serotype 2 (HAdV-2) (Human adenovirus 2)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus; Human mastadenovirus C
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 47 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03254-3|E1A-3_ADE02 Isoform of P03254 Isoform early E1A 6 kDa protein of Early E1A protein OS=Human adenovirus C serotype 2 OX=10515 PE=1 SV=1
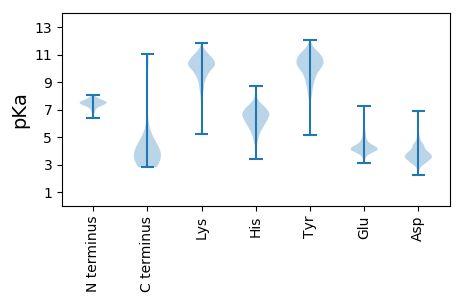
MM1 pKa = 7.5RR2 pKa = 11.84HH3 pKa = 6.51IICHH7 pKa = 5.93GGVITEE13 pKa = 4.22EE14 pKa = 4.15MAASLLDD21 pKa = 3.6QLIEE25 pKa = 4.19EE26 pKa = 4.48VLADD30 pKa = 3.85NLPPPSHH37 pKa = 7.16FEE39 pKa = 3.8PPTLHH44 pKa = 6.78EE45 pKa = 5.5LYY47 pKa = 10.58DD48 pKa = 4.45LDD50 pKa = 3.62VTAPEE55 pKa = 4.89DD56 pKa = 3.83PNEE59 pKa = 4.13EE60 pKa = 3.85AVSQIFPDD68 pKa = 3.76SVMLAVQEE76 pKa = 5.07GIDD79 pKa = 4.38LLTFPPAPGSPEE91 pKa = 3.95PPHH94 pKa = 7.1LSRR97 pKa = 11.84QPEE100 pKa = 4.1QPEE103 pKa = 3.86QRR105 pKa = 11.84ALGPVSMPNLVPEE118 pKa = 4.78VIDD121 pKa = 3.89LTCHH125 pKa = 5.71EE126 pKa = 5.16AGFPPSDD133 pKa = 4.54DD134 pKa = 3.73EE135 pKa = 5.56DD136 pKa = 4.3EE137 pKa = 4.68EE138 pKa = 5.74GPVSEE143 pKa = 5.29PEE145 pKa = 4.22PEE147 pKa = 4.37PEE149 pKa = 4.57PEE151 pKa = 4.34PEE153 pKa = 3.76PARR156 pKa = 11.84PTRR159 pKa = 11.84RR160 pKa = 11.84PKK162 pKa = 10.32LVPAILRR169 pKa = 11.84RR170 pKa = 11.84PTSPVSRR177 pKa = 11.84EE178 pKa = 4.32CNSSTDD184 pKa = 3.44SCDD187 pKa = 3.3SGPSNTPPEE196 pKa = 4.07IHH198 pKa = 6.71PVVPLCPIKK207 pKa = 10.4PVAVRR212 pKa = 11.84VGGRR216 pKa = 11.84RR217 pKa = 11.84QAVEE221 pKa = 4.48CIEE224 pKa = 4.76DD225 pKa = 3.84LLNEE229 pKa = 4.41SGQPLDD235 pKa = 4.51LSCKK239 pKa = 10.03RR240 pKa = 11.84PRR242 pKa = 11.84PP243 pKa = 3.69
MM1 pKa = 7.5RR2 pKa = 11.84HH3 pKa = 6.51IICHH7 pKa = 5.93GGVITEE13 pKa = 4.22EE14 pKa = 4.15MAASLLDD21 pKa = 3.6QLIEE25 pKa = 4.19EE26 pKa = 4.48VLADD30 pKa = 3.85NLPPPSHH37 pKa = 7.16FEE39 pKa = 3.8PPTLHH44 pKa = 6.78EE45 pKa = 5.5LYY47 pKa = 10.58DD48 pKa = 4.45LDD50 pKa = 3.62VTAPEE55 pKa = 4.89DD56 pKa = 3.83PNEE59 pKa = 4.13EE60 pKa = 3.85AVSQIFPDD68 pKa = 3.76SVMLAVQEE76 pKa = 5.07GIDD79 pKa = 4.38LLTFPPAPGSPEE91 pKa = 3.95PPHH94 pKa = 7.1LSRR97 pKa = 11.84QPEE100 pKa = 4.1QPEE103 pKa = 3.86QRR105 pKa = 11.84ALGPVSMPNLVPEE118 pKa = 4.78VIDD121 pKa = 3.89LTCHH125 pKa = 5.71EE126 pKa = 5.16AGFPPSDD133 pKa = 4.54DD134 pKa = 3.73EE135 pKa = 5.56DD136 pKa = 4.3EE137 pKa = 4.68EE138 pKa = 5.74GPVSEE143 pKa = 5.29PEE145 pKa = 4.22PEE147 pKa = 4.37PEE149 pKa = 4.57PEE151 pKa = 4.34PEE153 pKa = 3.76PARR156 pKa = 11.84PTRR159 pKa = 11.84RR160 pKa = 11.84PKK162 pKa = 10.32LVPAILRR169 pKa = 11.84RR170 pKa = 11.84PTSPVSRR177 pKa = 11.84EE178 pKa = 4.32CNSSTDD184 pKa = 3.44SCDD187 pKa = 3.3SGPSNTPPEE196 pKa = 4.07IHH198 pKa = 6.71PVVPLCPIKK207 pKa = 10.4PVAVRR212 pKa = 11.84VGGRR216 pKa = 11.84RR217 pKa = 11.84QAVEE221 pKa = 4.48CIEE224 pKa = 4.76DD225 pKa = 3.84LLNEE229 pKa = 4.41SGQPLDD235 pKa = 4.51LSCKK239 pKa = 10.03RR240 pKa = 11.84PRR242 pKa = 11.84PP243 pKa = 3.69
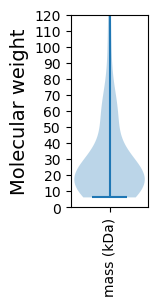
Molecular weight: 26.45 kDa
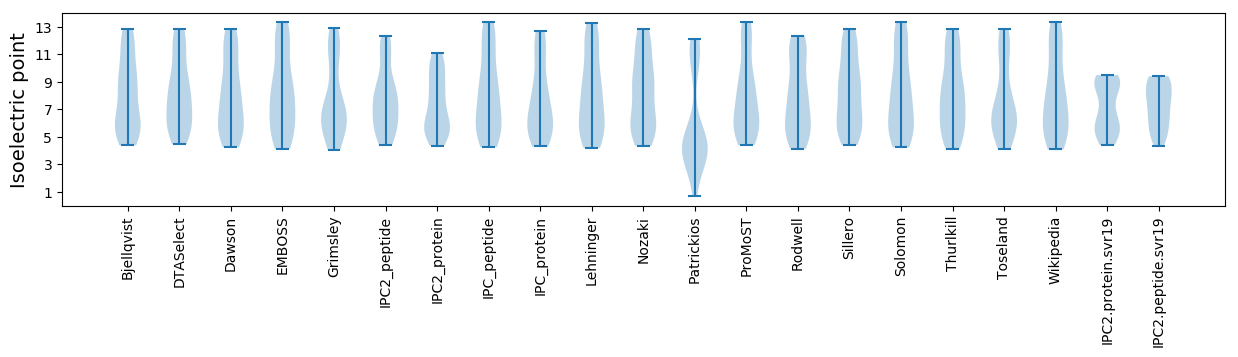
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P15133|E3RDA_ADE02 Pre-early 3 receptor internalization and degradation alpha protein OS=Human adenovirus C serotype 2 OX=10515 PE=1 SV=1
MM1 pKa = 7.77ALTCRR6 pKa = 11.84LRR8 pKa = 11.84FPVPGFRR15 pKa = 11.84GRR17 pKa = 11.84MHH19 pKa = 7.55RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GMAGHH27 pKa = 7.2GLTGGMRR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.79HH38 pKa = 5.82RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84ASHH45 pKa = 5.58RR46 pKa = 11.84RR47 pKa = 11.84MRR49 pKa = 11.84GGILPLLIPLIAAAIGAVPGIASVALQAQRR79 pKa = 11.84HH80 pKa = 4.41
MM1 pKa = 7.77ALTCRR6 pKa = 11.84LRR8 pKa = 11.84FPVPGFRR15 pKa = 11.84GRR17 pKa = 11.84MHH19 pKa = 7.55RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GMAGHH27 pKa = 7.2GLTGGMRR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.79HH38 pKa = 5.82RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84ASHH45 pKa = 5.58RR46 pKa = 11.84RR47 pKa = 11.84MRR49 pKa = 11.84GGILPLLIPLIAAAIGAVPGIASVALQAQRR79 pKa = 11.84HH80 pKa = 4.41
Molecular weight: 8.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13172 |
55 |
1198 |
280.3 |
31.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.518 ± 0.361 | 1.906 ± 0.225 |
4.866 ± 0.281 | 6.051 ± 0.418 |
3.356 ± 0.205 | 6.445 ± 0.29 |
2.353 ± 0.157 | 3.743 ± 0.196 |
3.477 ± 0.348 | 9.057 ± 0.367 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.467 ± 0.168 | 3.971 ± 0.431 |
7.653 ± 0.459 | 4.1 ± 0.223 |
8.381 ± 0.555 | 7.432 ± 0.481 |
5.975 ± 0.425 | 6.165 ± 0.274 |
1.283 ± 0.112 | 2.801 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
