
Firmicutes bacterium CAG:424
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
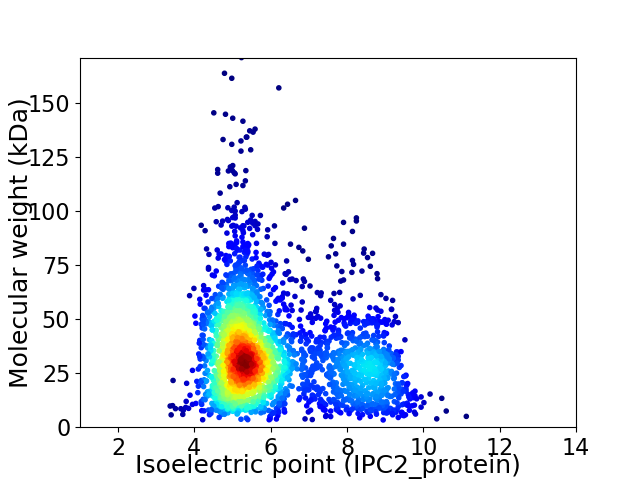
Virtual 2D-PAGE plot for 2835 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6R442|R6R442_9FIRM ATPases of the AAA+ class OS=Firmicutes bacterium CAG:424 OX=1263022 GN=BN652_00445 PE=4 SV=1
MM1 pKa = 7.69RR2 pKa = 11.84KK3 pKa = 7.9KK4 pKa = 8.85TAAIVTGFMITLGMLGGCGSAGDD27 pKa = 3.93STGNANASGEE37 pKa = 3.95NHH39 pKa = 6.95LNFGCYY45 pKa = 9.3VYY47 pKa = 10.05STSFDD52 pKa = 3.28PAAYY56 pKa = 10.06QNASWQGMRR65 pKa = 11.84WGITEE70 pKa = 3.76TLYY73 pKa = 11.01RR74 pKa = 11.84FNDD77 pKa = 3.77DD78 pKa = 3.13STIRR82 pKa = 11.84PWLSDD87 pKa = 3.31SYY89 pKa = 11.2EE90 pKa = 3.83VSEE93 pKa = 5.18DD94 pKa = 3.07HH95 pKa = 6.43KK96 pKa = 9.8TWTFHH101 pKa = 6.39IRR103 pKa = 11.84DD104 pKa = 3.53GVKK107 pKa = 10.22FSNGSPCDD115 pKa = 3.61AQAVADD121 pKa = 3.78SMNRR125 pKa = 11.84LFEE128 pKa = 4.35VCNSTEE134 pKa = 3.97YY135 pKa = 11.07SSTPRR140 pKa = 11.84QYY142 pKa = 10.95IDD144 pKa = 3.28MASIEE149 pKa = 4.74ADD151 pKa = 3.31PEE153 pKa = 4.5ANTVTIVTNTAYY165 pKa = 10.62ADD167 pKa = 3.64LRR169 pKa = 11.84GPLCFPFYY177 pKa = 10.51TIIDD181 pKa = 3.91VEE183 pKa = 4.44GDD185 pKa = 3.35LTDD188 pKa = 4.11AEE190 pKa = 4.74HH191 pKa = 6.96PGGYY195 pKa = 5.03TTSVIGTGPYY205 pKa = 10.4VLDD208 pKa = 4.55SFDD211 pKa = 4.19EE212 pKa = 4.32LSKK215 pKa = 11.17SGEE218 pKa = 3.97LVKK221 pKa = 11.1NEE223 pKa = 4.21NYY225 pKa = 9.96WNGDD229 pKa = 3.12VPYY232 pKa = 10.58DD233 pKa = 3.73SVTMMFIEE241 pKa = 5.78DD242 pKa = 3.95DD243 pKa = 3.73TTKK246 pKa = 11.32AMAIEE251 pKa = 4.65SGDD254 pKa = 3.24IDD256 pKa = 3.87LTEE259 pKa = 4.9NITTISDD266 pKa = 3.72LQKK269 pKa = 11.17LEE271 pKa = 4.35GDD273 pKa = 3.51DD274 pKa = 3.65AFYY277 pKa = 11.14VSKK280 pKa = 9.66KK281 pKa = 8.12TGVRR285 pKa = 11.84TGFALVNFDD294 pKa = 5.22GILGNDD300 pKa = 4.02TLRR303 pKa = 11.84QAVFMAIDD311 pKa = 4.02GQTLCDD317 pKa = 3.55VTVGGMYY324 pKa = 10.23NYY326 pKa = 10.2EE327 pKa = 4.41PGVLPDD333 pKa = 3.46TLAYY337 pKa = 10.43DD338 pKa = 3.63SEE340 pKa = 4.84KK341 pKa = 10.85LVNPYY346 pKa = 10.03TYY348 pKa = 10.21DD349 pKa = 3.26QEE351 pKa = 4.93AAIQLLDD358 pKa = 4.07DD359 pKa = 3.98NGIRR363 pKa = 11.84DD364 pKa = 3.75TDD366 pKa = 3.42GDD368 pKa = 4.38GIRR371 pKa = 11.84EE372 pKa = 3.9LDD374 pKa = 3.57GEE376 pKa = 4.51NIVLEE381 pKa = 3.81FDD383 pKa = 3.61TYY385 pKa = 11.57KK386 pKa = 10.69NRR388 pKa = 11.84CLSDD392 pKa = 3.47FAEE395 pKa = 5.98GIQSQLAAIGIGCNVNVLDD414 pKa = 6.28SDD416 pKa = 4.4TLWMEE421 pKa = 4.18MQEE424 pKa = 4.21GDD426 pKa = 3.68YY427 pKa = 11.32DD428 pKa = 4.16LSNSNWITVGTGDD441 pKa = 4.85PEE443 pKa = 4.79AFLLNWYY450 pKa = 10.26GDD452 pKa = 3.26GGEE455 pKa = 4.39NFFTEE460 pKa = 4.79DD461 pKa = 3.06NTDD464 pKa = 2.85ATNYY468 pKa = 9.44CRR470 pKa = 11.84YY471 pKa = 10.76DD472 pKa = 3.38NEE474 pKa = 4.72EE475 pKa = 3.92YY476 pKa = 11.02NKK478 pKa = 10.61LYY480 pKa = 10.58EE481 pKa = 3.99QFKK484 pKa = 10.85ASMDD488 pKa = 3.47EE489 pKa = 4.16EE490 pKa = 4.19EE491 pKa = 5.41RR492 pKa = 11.84KK493 pKa = 10.63DD494 pKa = 4.54LVIQMEE500 pKa = 4.14QVLIDD505 pKa = 3.75DD506 pKa = 4.6CAVLVHH512 pKa = 6.61GYY514 pKa = 10.14YY515 pKa = 10.43NSTMISNKK523 pKa = 9.62EE524 pKa = 3.8KK525 pKa = 10.69VAGAEE530 pKa = 3.95IPAFDD535 pKa = 4.73YY536 pKa = 11.24YY537 pKa = 10.71WLTTDD542 pKa = 4.56IKK544 pKa = 10.57PAEE547 pKa = 4.04
MM1 pKa = 7.69RR2 pKa = 11.84KK3 pKa = 7.9KK4 pKa = 8.85TAAIVTGFMITLGMLGGCGSAGDD27 pKa = 3.93STGNANASGEE37 pKa = 3.95NHH39 pKa = 6.95LNFGCYY45 pKa = 9.3VYY47 pKa = 10.05STSFDD52 pKa = 3.28PAAYY56 pKa = 10.06QNASWQGMRR65 pKa = 11.84WGITEE70 pKa = 3.76TLYY73 pKa = 11.01RR74 pKa = 11.84FNDD77 pKa = 3.77DD78 pKa = 3.13STIRR82 pKa = 11.84PWLSDD87 pKa = 3.31SYY89 pKa = 11.2EE90 pKa = 3.83VSEE93 pKa = 5.18DD94 pKa = 3.07HH95 pKa = 6.43KK96 pKa = 9.8TWTFHH101 pKa = 6.39IRR103 pKa = 11.84DD104 pKa = 3.53GVKK107 pKa = 10.22FSNGSPCDD115 pKa = 3.61AQAVADD121 pKa = 3.78SMNRR125 pKa = 11.84LFEE128 pKa = 4.35VCNSTEE134 pKa = 3.97YY135 pKa = 11.07SSTPRR140 pKa = 11.84QYY142 pKa = 10.95IDD144 pKa = 3.28MASIEE149 pKa = 4.74ADD151 pKa = 3.31PEE153 pKa = 4.5ANTVTIVTNTAYY165 pKa = 10.62ADD167 pKa = 3.64LRR169 pKa = 11.84GPLCFPFYY177 pKa = 10.51TIIDD181 pKa = 3.91VEE183 pKa = 4.44GDD185 pKa = 3.35LTDD188 pKa = 4.11AEE190 pKa = 4.74HH191 pKa = 6.96PGGYY195 pKa = 5.03TTSVIGTGPYY205 pKa = 10.4VLDD208 pKa = 4.55SFDD211 pKa = 4.19EE212 pKa = 4.32LSKK215 pKa = 11.17SGEE218 pKa = 3.97LVKK221 pKa = 11.1NEE223 pKa = 4.21NYY225 pKa = 9.96WNGDD229 pKa = 3.12VPYY232 pKa = 10.58DD233 pKa = 3.73SVTMMFIEE241 pKa = 5.78DD242 pKa = 3.95DD243 pKa = 3.73TTKK246 pKa = 11.32AMAIEE251 pKa = 4.65SGDD254 pKa = 3.24IDD256 pKa = 3.87LTEE259 pKa = 4.9NITTISDD266 pKa = 3.72LQKK269 pKa = 11.17LEE271 pKa = 4.35GDD273 pKa = 3.51DD274 pKa = 3.65AFYY277 pKa = 11.14VSKK280 pKa = 9.66KK281 pKa = 8.12TGVRR285 pKa = 11.84TGFALVNFDD294 pKa = 5.22GILGNDD300 pKa = 4.02TLRR303 pKa = 11.84QAVFMAIDD311 pKa = 4.02GQTLCDD317 pKa = 3.55VTVGGMYY324 pKa = 10.23NYY326 pKa = 10.2EE327 pKa = 4.41PGVLPDD333 pKa = 3.46TLAYY337 pKa = 10.43DD338 pKa = 3.63SEE340 pKa = 4.84KK341 pKa = 10.85LVNPYY346 pKa = 10.03TYY348 pKa = 10.21DD349 pKa = 3.26QEE351 pKa = 4.93AAIQLLDD358 pKa = 4.07DD359 pKa = 3.98NGIRR363 pKa = 11.84DD364 pKa = 3.75TDD366 pKa = 3.42GDD368 pKa = 4.38GIRR371 pKa = 11.84EE372 pKa = 3.9LDD374 pKa = 3.57GEE376 pKa = 4.51NIVLEE381 pKa = 3.81FDD383 pKa = 3.61TYY385 pKa = 11.57KK386 pKa = 10.69NRR388 pKa = 11.84CLSDD392 pKa = 3.47FAEE395 pKa = 5.98GIQSQLAAIGIGCNVNVLDD414 pKa = 6.28SDD416 pKa = 4.4TLWMEE421 pKa = 4.18MQEE424 pKa = 4.21GDD426 pKa = 3.68YY427 pKa = 11.32DD428 pKa = 4.16LSNSNWITVGTGDD441 pKa = 4.85PEE443 pKa = 4.79AFLLNWYY450 pKa = 10.26GDD452 pKa = 3.26GGEE455 pKa = 4.39NFFTEE460 pKa = 4.79DD461 pKa = 3.06NTDD464 pKa = 2.85ATNYY468 pKa = 9.44CRR470 pKa = 11.84YY471 pKa = 10.76DD472 pKa = 3.38NEE474 pKa = 4.72EE475 pKa = 3.92YY476 pKa = 11.02NKK478 pKa = 10.61LYY480 pKa = 10.58EE481 pKa = 3.99QFKK484 pKa = 10.85ASMDD488 pKa = 3.47EE489 pKa = 4.16EE490 pKa = 4.19EE491 pKa = 5.41RR492 pKa = 11.84KK493 pKa = 10.63DD494 pKa = 4.54LVIQMEE500 pKa = 4.14QVLIDD505 pKa = 3.75DD506 pKa = 4.6CAVLVHH512 pKa = 6.61GYY514 pKa = 10.14YY515 pKa = 10.43NSTMISNKK523 pKa = 9.62EE524 pKa = 3.8KK525 pKa = 10.69VAGAEE530 pKa = 3.95IPAFDD535 pKa = 4.73YY536 pKa = 11.24YY537 pKa = 10.71WLTTDD542 pKa = 4.56IKK544 pKa = 10.57PAEE547 pKa = 4.04
Molecular weight: 60.77 kDa
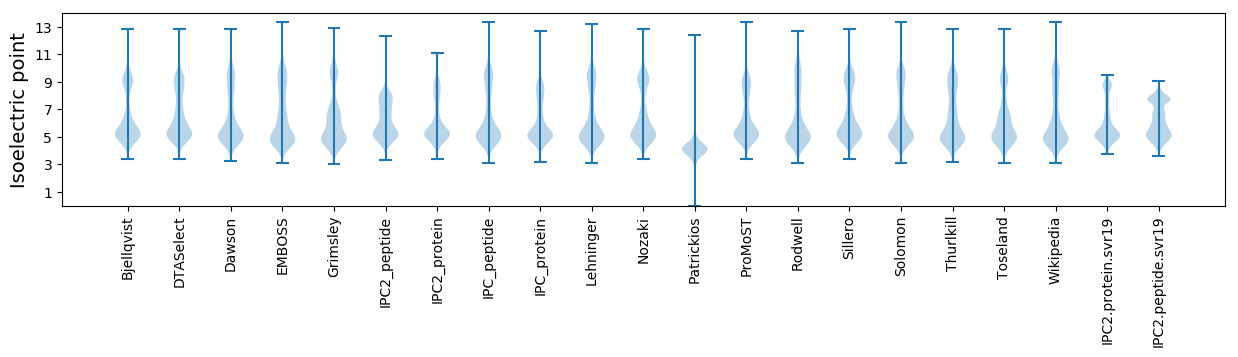
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6RNI8|R6RNI8_9FIRM Abhydrolase_3 domain-containing protein OS=Firmicutes bacterium CAG:424 OX=1263022 GN=BN652_01611 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
903456 |
29 |
1505 |
318.7 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.085 ± 0.045 | 1.551 ± 0.02 |
4.976 ± 0.035 | 8.381 ± 0.054 |
4.289 ± 0.033 | 7.151 ± 0.049 |
1.804 ± 0.022 | 7.092 ± 0.041 |
7.144 ± 0.039 | 9.387 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.247 ± 0.021 | 4.076 ± 0.029 |
3.351 ± 0.028 | 3.688 ± 0.032 |
4.201 ± 0.032 | 5.458 ± 0.031 |
5.182 ± 0.028 | 6.803 ± 0.037 |
0.986 ± 0.017 | 4.147 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
