
Luteimonas cucumeris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Luteimonas
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
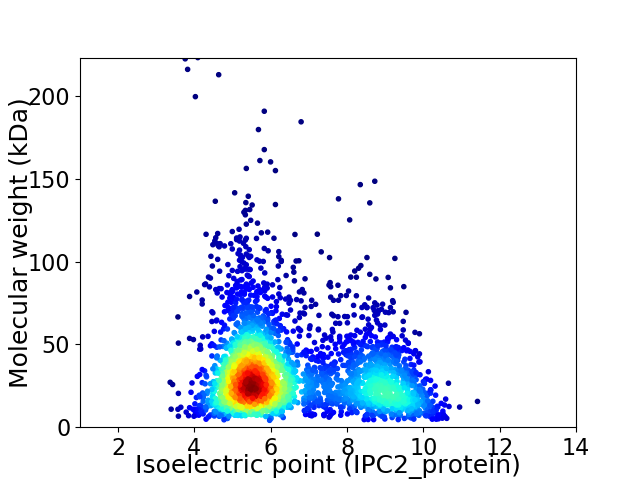
Virtual 2D-PAGE plot for 3213 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A562KUE3|A0A562KUE3_9GAMM Uncharacterized protein DUF4177 OS=Luteimonas cucumeris OX=985012 GN=IP90_03228 PE=4 SV=1
MM1 pKa = 7.35NKK3 pKa = 9.03VFSLVWSRR11 pKa = 11.84SLQALVVASEE21 pKa = 4.13LASANGRR28 pKa = 11.84KK29 pKa = 9.61AGRR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84LAHH38 pKa = 6.19AAFGVVVLSASFSLAPLAQAAEE60 pKa = 4.47SGKK63 pKa = 8.05PTCTVNLPGQAGVAVDD79 pKa = 4.46CAQLQQIVATAQRR92 pKa = 11.84AGDD95 pKa = 3.66PRR97 pKa = 11.84KK98 pKa = 9.98GRR100 pKa = 11.84KK101 pKa = 8.66ARR103 pKa = 11.84PVVDD107 pKa = 5.12SFDD110 pKa = 3.63SDD112 pKa = 3.84YY113 pKa = 11.44FKK115 pKa = 11.52ANGTGDD121 pKa = 3.88GDD123 pKa = 3.83DD124 pKa = 4.95ASAQGEE130 pKa = 4.21GAIAIGSGAQTRR142 pKa = 11.84PWVSDD147 pKa = 3.22EE148 pKa = 4.27LGEE151 pKa = 4.35SFEE154 pKa = 4.92AEE156 pKa = 3.79GAVAIGRR163 pKa = 11.84QSVAEE168 pKa = 4.25GWEE171 pKa = 4.11ATALGANSFASSNSATAVGGEE192 pKa = 4.19SGAFGIGSSAFGQGSKK208 pKa = 11.0AFGHH212 pKa = 6.93AATALGQGAMTFGFGNTAVGASAMASAEE240 pKa = 4.02QAATALGALSEE251 pKa = 4.16AWSNGSTAVGGMAIAAGEE269 pKa = 4.22GSLAAGFDD277 pKa = 3.79SQAGGFEE284 pKa = 4.42SVALGGHH291 pKa = 4.75TRR293 pKa = 11.84VGNGEE298 pKa = 4.05AGVGVGANAVALGNGAVGVGNNSLANGSNTVAMGRR333 pKa = 11.84NAAANSEE340 pKa = 4.1GALAIGGSEE349 pKa = 4.03WTDD352 pKa = 3.02HH353 pKa = 7.32DD354 pKa = 5.0GDD356 pKa = 3.89GMPDD360 pKa = 3.08VGEE363 pKa = 4.07ATLAEE368 pKa = 4.42GQNAVAIGGLAGASGDD384 pKa = 3.45NAQAIGHH391 pKa = 5.58QANALAEE398 pKa = 4.2NATATGAMSMAAGEE412 pKa = 4.24NSTATGASSVANGIDD427 pKa = 3.2SSAYY431 pKa = 9.48GAQSQADD438 pKa = 4.12GDD440 pKa = 3.94HH441 pKa = 5.95STAIGSGSLALGVGSSAVGSGANAFGEE468 pKa = 4.29YY469 pKa = 10.14SVANGYY475 pKa = 10.26GSAAEE480 pKa = 4.33GADD483 pKa = 3.15AVAVGSNSAASGSGASALGAAAQAVGTQSAAMGFNSQALEE523 pKa = 4.03MATTAVGGQTVANQWFATAIGYY545 pKa = 8.85GSQATGYY552 pKa = 10.27SSVALGEE559 pKa = 4.31NVLATGEE566 pKa = 3.93MSIAIGNSWAGMMQTQSNGNMSVAIGSAAATWDD599 pKa = 3.57NYY601 pKa = 9.51GVAVGSLAGANTEE614 pKa = 4.01YY615 pKa = 9.6STSVGAMSAAIGQQSTALGAAAMAEE640 pKa = 4.19ADD642 pKa = 3.75DD643 pKa = 4.4SVAIGYY649 pKa = 8.99YY650 pKa = 9.87SHH652 pKa = 7.34ASGANSVALGNEE664 pKa = 4.43SVASRR669 pKa = 11.84DD670 pKa = 3.43NTISVGKK677 pKa = 10.17AGSEE681 pKa = 3.76RR682 pKa = 11.84QITNVAAGTEE692 pKa = 4.09ATDD695 pKa = 4.07AVNKK699 pKa = 10.14AQLDD703 pKa = 4.02EE704 pKa = 4.44VATGVADD711 pKa = 3.93ANHH714 pKa = 5.87YY715 pKa = 9.26FAANGAADD723 pKa = 4.09GSDD726 pKa = 3.4DD727 pKa = 3.8AMASGDD733 pKa = 3.74HH734 pKa = 6.32AVATGAYY741 pKa = 8.86AVANGIEE748 pKa = 4.2SSAYY752 pKa = 9.91GYY754 pKa = 10.39QSQADD759 pKa = 3.72GDD761 pKa = 4.06YY762 pKa = 10.72SQAIGSGSLAMGVGTSAVGSGANAIGNGASAFGAGAVAFLDD803 pKa = 4.06GATAVGSNAQTAGLNTVAVGNGAAASATGAVAIGGQATNGLGQPILADD851 pKa = 3.79GGTSEE856 pKa = 5.03PLGAAQAAEE865 pKa = 4.25LSTAVGASTYY875 pKa = 10.17ATGNWSAAYY884 pKa = 9.7GVGAWSAGSQSTAVGALAIAYY905 pKa = 8.21GDD907 pKa = 3.68SSSAFGTNTMASGQSSTAIGQSAWALGDD935 pKa = 3.42YY936 pKa = 10.57SVAIGVAANTSGSNTVAIGNSATVFDD962 pKa = 4.38YY963 pKa = 9.75STWSNVSDD971 pKa = 3.65AVALGSASMASADD984 pKa = 3.53HH985 pKa = 6.65AVALGSNSHH994 pKa = 6.16AMADD998 pKa = 3.44NSVALGQGSLADD1010 pKa = 3.9RR1011 pKa = 11.84ANTVSVGAAEE1021 pKa = 4.17AWTDD1025 pKa = 3.7LLGTSHH1031 pKa = 6.68EE1032 pKa = 4.76AIDD1035 pKa = 3.76RR1036 pKa = 11.84QITNVAAGTEE1046 pKa = 4.09ATDD1049 pKa = 4.99AVNKK1053 pKa = 10.24DD1054 pKa = 3.52QLDD1057 pKa = 3.67DD1058 pKa = 3.38ATRR1061 pKa = 11.84YY1062 pKa = 10.34LEE1064 pKa = 4.59ANGAGDD1070 pKa = 3.92GSDD1073 pKa = 3.77DD1074 pKa = 4.17AVATGQYY1081 pKa = 8.6ATATGANSIANGAEE1095 pKa = 3.83SSVYY1099 pKa = 10.3GYY1101 pKa = 10.58QSQADD1106 pKa = 3.72GDD1108 pKa = 4.17YY1109 pKa = 9.56ATAIGSSSYY1118 pKa = 11.31AGALGANAIGTGATALGEE1136 pKa = 4.11TSNAIGYY1143 pKa = 9.85GSVATGLDD1151 pKa = 3.29AVAVGTSSNASGVNALAMGAGSSASADD1178 pKa = 3.11VSVAINTGSVAGLWGVAPGVNAAGGVGIGAAAVVGANADD1217 pKa = 3.46AGVALGLSATVVEE1230 pKa = 4.89GAYY1233 pKa = 10.04QGVAIGMGSYY1243 pKa = 10.46VGDD1246 pKa = 3.68AQGIAIGGFGARR1258 pKa = 11.84FIDD1261 pKa = 4.42GSIEE1265 pKa = 3.91LRR1267 pKa = 11.84PTVAMGTGAVAIGLGSEE1284 pKa = 4.42TFNDD1288 pKa = 3.36HH1289 pKa = 5.65ATALGAFSTVTAQNAVAIGANSLADD1314 pKa = 4.03RR1315 pKa = 11.84DD1316 pKa = 3.96NTLSVGYY1323 pKa = 9.92ADD1325 pKa = 4.8HH1326 pKa = 6.95EE1327 pKa = 4.25RR1328 pKa = 11.84QITHH1332 pKa = 5.84VAAGTEE1338 pKa = 4.03ATDD1341 pKa = 4.99AVNKK1345 pKa = 10.24DD1346 pKa = 3.52QLDD1349 pKa = 3.68DD1350 pKa = 3.5ATRR1353 pKa = 11.84YY1354 pKa = 8.88LASNGAGDD1362 pKa = 4.17GSDD1365 pKa = 3.84DD1366 pKa = 4.07AVATGDD1372 pKa = 3.48QAVATGASAVANGVGSSAYY1391 pKa = 9.97GSQGYY1396 pKa = 10.99ADD1398 pKa = 4.52GDD1400 pKa = 3.85YY1401 pKa = 10.04STAVGSGSLALGAGASAFGSGANALGEE1428 pKa = 4.12NSTALGTGALASGFGAYY1445 pKa = 10.5AMGNASNAAARR1456 pKa = 11.84NSLALGGMASVDD1468 pKa = 3.4QSSALGTAIGYY1479 pKa = 7.92GAYY1482 pKa = 9.54IGANSSAAIAMGFAAGVGDD1501 pKa = 3.88GASNSISMGNLSYY1514 pKa = 11.33ADD1516 pKa = 3.8SPYY1519 pKa = 10.31TIAIGSMFAYY1529 pKa = 10.23VNHH1532 pKa = 7.39DD1533 pKa = 3.55GGLGLATTGASGWGAVAFGPGAQALGDD1560 pKa = 3.74YY1561 pKa = 9.55TLALGPAASVTAANAVALGGLSIADD1586 pKa = 4.05RR1587 pKa = 11.84EE1588 pKa = 4.45NTVSVGAADD1597 pKa = 4.1HH1598 pKa = 6.29EE1599 pKa = 4.32RR1600 pKa = 11.84QIVNVAAGTEE1610 pKa = 4.0ATDD1613 pKa = 3.73AVNVAQLDD1621 pKa = 3.73DD1622 pKa = 3.47ATRR1625 pKa = 11.84YY1626 pKa = 9.3VAVNGAGDD1634 pKa = 3.82GSDD1637 pKa = 3.74DD1638 pKa = 4.31AVATGEE1644 pKa = 4.09NATATGASSVANGVEE1659 pKa = 4.09SSAYY1663 pKa = 10.03GSQSYY1668 pKa = 11.5ADD1670 pKa = 4.09ADD1672 pKa = 3.75YY1673 pKa = 10.0STALGSNSLALGIGASAIGSGANALGTNSVANGYY1707 pKa = 10.91ASVASGEE1714 pKa = 4.03LSIAQGSEE1722 pKa = 3.84SQALGEE1728 pKa = 4.03ASIALGTQSLAEE1740 pKa = 5.08GIDD1743 pKa = 3.92SIGVGAGSYY1752 pKa = 10.67ASVYY1756 pKa = 8.0GTAVGTLSQAGMYY1769 pKa = 10.44GSTALGHH1776 pKa = 6.2SATANNDD1783 pKa = 2.86FALALGAVSTSLAVGGIAIGTEE1805 pKa = 4.22SYY1807 pKa = 11.57VDD1809 pKa = 3.32ANAYY1813 pKa = 9.37QGIAIGSVVSAIGHH1827 pKa = 5.92RR1828 pKa = 11.84SIAIGSQWTTAGGDD1842 pKa = 3.52SAVAIGGTMACSGICIEE1859 pKa = 4.74QNNAAWGANSVALGAGTQAQADD1881 pKa = 4.18GATALGAGANAGGIGSVAAGQFAQAWSDD1909 pKa = 3.65DD1910 pKa = 3.47AVALGNTSQAIAAGSVALGAYY1931 pKa = 9.42SVADD1935 pKa = 3.81RR1936 pKa = 11.84DD1937 pKa = 3.99NTVSVGDD1944 pKa = 4.08AGTGLTRR1951 pKa = 11.84QITNVAAGTEE1961 pKa = 4.18ANDD1964 pKa = 4.01AVNKK1968 pKa = 9.86GQLDD1972 pKa = 3.96EE1973 pKa = 5.34LSDD1976 pKa = 4.3SIGDD1980 pKa = 3.62ANYY1983 pKa = 10.73YY1984 pKa = 9.66FAADD1988 pKa = 3.85GAGDD1992 pKa = 3.88GSDD1995 pKa = 3.25AAVASGVGSVAAGANSVASGTGTVAYY2021 pKa = 10.16GRR2023 pKa = 11.84GALASGEE2030 pKa = 3.96QAMAIGDD2037 pKa = 4.11GAQADD2042 pKa = 4.32YY2043 pKa = 9.88FYY2045 pKa = 11.75AMAIGAGAVSSSWASMAIGRR2065 pKa = 11.84NAIASGPRR2073 pKa = 11.84AVAIGEE2079 pKa = 4.1NAQAMDD2085 pKa = 4.11NGNVALGWASQSVGMFSAALGGSSAAIGWNTLAAGFGAWVLGNNATAVGAGAGSFYY2141 pKa = 10.66TVDD2144 pKa = 3.97EE2145 pKa = 4.24ITTVGSNARR2154 pKa = 11.84ADD2156 pKa = 3.79SFRR2159 pKa = 11.84GTVVGAGSVAAEE2171 pKa = 4.09WAPEE2175 pKa = 4.0STIVGANAFSFDD2187 pKa = 4.17SNGTAIGEE2195 pKa = 4.32SSQVSYY2201 pKa = 11.02GAANAVALGSGSHH2214 pKa = 7.05ADD2216 pKa = 3.66RR2217 pKa = 11.84ANTVSVGASGPWTDD2231 pKa = 3.43GFGVEE2236 pKa = 4.82HH2237 pKa = 7.46PALQRR2242 pKa = 11.84QITNVAAGTEE2252 pKa = 3.99ASDD2255 pKa = 4.44AVNKK2259 pKa = 10.29AQLDD2263 pKa = 3.88EE2264 pKa = 4.15VAAAGEE2270 pKa = 4.13ATGKK2274 pKa = 10.35FFF2276 pKa = 5.64
MM1 pKa = 7.35NKK3 pKa = 9.03VFSLVWSRR11 pKa = 11.84SLQALVVASEE21 pKa = 4.13LASANGRR28 pKa = 11.84KK29 pKa = 9.61AGRR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84LAHH38 pKa = 6.19AAFGVVVLSASFSLAPLAQAAEE60 pKa = 4.47SGKK63 pKa = 8.05PTCTVNLPGQAGVAVDD79 pKa = 4.46CAQLQQIVATAQRR92 pKa = 11.84AGDD95 pKa = 3.66PRR97 pKa = 11.84KK98 pKa = 9.98GRR100 pKa = 11.84KK101 pKa = 8.66ARR103 pKa = 11.84PVVDD107 pKa = 5.12SFDD110 pKa = 3.63SDD112 pKa = 3.84YY113 pKa = 11.44FKK115 pKa = 11.52ANGTGDD121 pKa = 3.88GDD123 pKa = 3.83DD124 pKa = 4.95ASAQGEE130 pKa = 4.21GAIAIGSGAQTRR142 pKa = 11.84PWVSDD147 pKa = 3.22EE148 pKa = 4.27LGEE151 pKa = 4.35SFEE154 pKa = 4.92AEE156 pKa = 3.79GAVAIGRR163 pKa = 11.84QSVAEE168 pKa = 4.25GWEE171 pKa = 4.11ATALGANSFASSNSATAVGGEE192 pKa = 4.19SGAFGIGSSAFGQGSKK208 pKa = 11.0AFGHH212 pKa = 6.93AATALGQGAMTFGFGNTAVGASAMASAEE240 pKa = 4.02QAATALGALSEE251 pKa = 4.16AWSNGSTAVGGMAIAAGEE269 pKa = 4.22GSLAAGFDD277 pKa = 3.79SQAGGFEE284 pKa = 4.42SVALGGHH291 pKa = 4.75TRR293 pKa = 11.84VGNGEE298 pKa = 4.05AGVGVGANAVALGNGAVGVGNNSLANGSNTVAMGRR333 pKa = 11.84NAAANSEE340 pKa = 4.1GALAIGGSEE349 pKa = 4.03WTDD352 pKa = 3.02HH353 pKa = 7.32DD354 pKa = 5.0GDD356 pKa = 3.89GMPDD360 pKa = 3.08VGEE363 pKa = 4.07ATLAEE368 pKa = 4.42GQNAVAIGGLAGASGDD384 pKa = 3.45NAQAIGHH391 pKa = 5.58QANALAEE398 pKa = 4.2NATATGAMSMAAGEE412 pKa = 4.24NSTATGASSVANGIDD427 pKa = 3.2SSAYY431 pKa = 9.48GAQSQADD438 pKa = 4.12GDD440 pKa = 3.94HH441 pKa = 5.95STAIGSGSLALGVGSSAVGSGANAFGEE468 pKa = 4.29YY469 pKa = 10.14SVANGYY475 pKa = 10.26GSAAEE480 pKa = 4.33GADD483 pKa = 3.15AVAVGSNSAASGSGASALGAAAQAVGTQSAAMGFNSQALEE523 pKa = 4.03MATTAVGGQTVANQWFATAIGYY545 pKa = 8.85GSQATGYY552 pKa = 10.27SSVALGEE559 pKa = 4.31NVLATGEE566 pKa = 3.93MSIAIGNSWAGMMQTQSNGNMSVAIGSAAATWDD599 pKa = 3.57NYY601 pKa = 9.51GVAVGSLAGANTEE614 pKa = 4.01YY615 pKa = 9.6STSVGAMSAAIGQQSTALGAAAMAEE640 pKa = 4.19ADD642 pKa = 3.75DD643 pKa = 4.4SVAIGYY649 pKa = 8.99YY650 pKa = 9.87SHH652 pKa = 7.34ASGANSVALGNEE664 pKa = 4.43SVASRR669 pKa = 11.84DD670 pKa = 3.43NTISVGKK677 pKa = 10.17AGSEE681 pKa = 3.76RR682 pKa = 11.84QITNVAAGTEE692 pKa = 4.09ATDD695 pKa = 4.07AVNKK699 pKa = 10.14AQLDD703 pKa = 4.02EE704 pKa = 4.44VATGVADD711 pKa = 3.93ANHH714 pKa = 5.87YY715 pKa = 9.26FAANGAADD723 pKa = 4.09GSDD726 pKa = 3.4DD727 pKa = 3.8AMASGDD733 pKa = 3.74HH734 pKa = 6.32AVATGAYY741 pKa = 8.86AVANGIEE748 pKa = 4.2SSAYY752 pKa = 9.91GYY754 pKa = 10.39QSQADD759 pKa = 3.72GDD761 pKa = 4.06YY762 pKa = 10.72SQAIGSGSLAMGVGTSAVGSGANAIGNGASAFGAGAVAFLDD803 pKa = 4.06GATAVGSNAQTAGLNTVAVGNGAAASATGAVAIGGQATNGLGQPILADD851 pKa = 3.79GGTSEE856 pKa = 5.03PLGAAQAAEE865 pKa = 4.25LSTAVGASTYY875 pKa = 10.17ATGNWSAAYY884 pKa = 9.7GVGAWSAGSQSTAVGALAIAYY905 pKa = 8.21GDD907 pKa = 3.68SSSAFGTNTMASGQSSTAIGQSAWALGDD935 pKa = 3.42YY936 pKa = 10.57SVAIGVAANTSGSNTVAIGNSATVFDD962 pKa = 4.38YY963 pKa = 9.75STWSNVSDD971 pKa = 3.65AVALGSASMASADD984 pKa = 3.53HH985 pKa = 6.65AVALGSNSHH994 pKa = 6.16AMADD998 pKa = 3.44NSVALGQGSLADD1010 pKa = 3.9RR1011 pKa = 11.84ANTVSVGAAEE1021 pKa = 4.17AWTDD1025 pKa = 3.7LLGTSHH1031 pKa = 6.68EE1032 pKa = 4.76AIDD1035 pKa = 3.76RR1036 pKa = 11.84QITNVAAGTEE1046 pKa = 4.09ATDD1049 pKa = 4.99AVNKK1053 pKa = 10.24DD1054 pKa = 3.52QLDD1057 pKa = 3.67DD1058 pKa = 3.38ATRR1061 pKa = 11.84YY1062 pKa = 10.34LEE1064 pKa = 4.59ANGAGDD1070 pKa = 3.92GSDD1073 pKa = 3.77DD1074 pKa = 4.17AVATGQYY1081 pKa = 8.6ATATGANSIANGAEE1095 pKa = 3.83SSVYY1099 pKa = 10.3GYY1101 pKa = 10.58QSQADD1106 pKa = 3.72GDD1108 pKa = 4.17YY1109 pKa = 9.56ATAIGSSSYY1118 pKa = 11.31AGALGANAIGTGATALGEE1136 pKa = 4.11TSNAIGYY1143 pKa = 9.85GSVATGLDD1151 pKa = 3.29AVAVGTSSNASGVNALAMGAGSSASADD1178 pKa = 3.11VSVAINTGSVAGLWGVAPGVNAAGGVGIGAAAVVGANADD1217 pKa = 3.46AGVALGLSATVVEE1230 pKa = 4.89GAYY1233 pKa = 10.04QGVAIGMGSYY1243 pKa = 10.46VGDD1246 pKa = 3.68AQGIAIGGFGARR1258 pKa = 11.84FIDD1261 pKa = 4.42GSIEE1265 pKa = 3.91LRR1267 pKa = 11.84PTVAMGTGAVAIGLGSEE1284 pKa = 4.42TFNDD1288 pKa = 3.36HH1289 pKa = 5.65ATALGAFSTVTAQNAVAIGANSLADD1314 pKa = 4.03RR1315 pKa = 11.84DD1316 pKa = 3.96NTLSVGYY1323 pKa = 9.92ADD1325 pKa = 4.8HH1326 pKa = 6.95EE1327 pKa = 4.25RR1328 pKa = 11.84QITHH1332 pKa = 5.84VAAGTEE1338 pKa = 4.03ATDD1341 pKa = 4.99AVNKK1345 pKa = 10.24DD1346 pKa = 3.52QLDD1349 pKa = 3.68DD1350 pKa = 3.5ATRR1353 pKa = 11.84YY1354 pKa = 8.88LASNGAGDD1362 pKa = 4.17GSDD1365 pKa = 3.84DD1366 pKa = 4.07AVATGDD1372 pKa = 3.48QAVATGASAVANGVGSSAYY1391 pKa = 9.97GSQGYY1396 pKa = 10.99ADD1398 pKa = 4.52GDD1400 pKa = 3.85YY1401 pKa = 10.04STAVGSGSLALGAGASAFGSGANALGEE1428 pKa = 4.12NSTALGTGALASGFGAYY1445 pKa = 10.5AMGNASNAAARR1456 pKa = 11.84NSLALGGMASVDD1468 pKa = 3.4QSSALGTAIGYY1479 pKa = 7.92GAYY1482 pKa = 9.54IGANSSAAIAMGFAAGVGDD1501 pKa = 3.88GASNSISMGNLSYY1514 pKa = 11.33ADD1516 pKa = 3.8SPYY1519 pKa = 10.31TIAIGSMFAYY1529 pKa = 10.23VNHH1532 pKa = 7.39DD1533 pKa = 3.55GGLGLATTGASGWGAVAFGPGAQALGDD1560 pKa = 3.74YY1561 pKa = 9.55TLALGPAASVTAANAVALGGLSIADD1586 pKa = 4.05RR1587 pKa = 11.84EE1588 pKa = 4.45NTVSVGAADD1597 pKa = 4.1HH1598 pKa = 6.29EE1599 pKa = 4.32RR1600 pKa = 11.84QIVNVAAGTEE1610 pKa = 4.0ATDD1613 pKa = 3.73AVNVAQLDD1621 pKa = 3.73DD1622 pKa = 3.47ATRR1625 pKa = 11.84YY1626 pKa = 9.3VAVNGAGDD1634 pKa = 3.82GSDD1637 pKa = 3.74DD1638 pKa = 4.31AVATGEE1644 pKa = 4.09NATATGASSVANGVEE1659 pKa = 4.09SSAYY1663 pKa = 10.03GSQSYY1668 pKa = 11.5ADD1670 pKa = 4.09ADD1672 pKa = 3.75YY1673 pKa = 10.0STALGSNSLALGIGASAIGSGANALGTNSVANGYY1707 pKa = 10.91ASVASGEE1714 pKa = 4.03LSIAQGSEE1722 pKa = 3.84SQALGEE1728 pKa = 4.03ASIALGTQSLAEE1740 pKa = 5.08GIDD1743 pKa = 3.92SIGVGAGSYY1752 pKa = 10.67ASVYY1756 pKa = 8.0GTAVGTLSQAGMYY1769 pKa = 10.44GSTALGHH1776 pKa = 6.2SATANNDD1783 pKa = 2.86FALALGAVSTSLAVGGIAIGTEE1805 pKa = 4.22SYY1807 pKa = 11.57VDD1809 pKa = 3.32ANAYY1813 pKa = 9.37QGIAIGSVVSAIGHH1827 pKa = 5.92RR1828 pKa = 11.84SIAIGSQWTTAGGDD1842 pKa = 3.52SAVAIGGTMACSGICIEE1859 pKa = 4.74QNNAAWGANSVALGAGTQAQADD1881 pKa = 4.18GATALGAGANAGGIGSVAAGQFAQAWSDD1909 pKa = 3.65DD1910 pKa = 3.47AVALGNTSQAIAAGSVALGAYY1931 pKa = 9.42SVADD1935 pKa = 3.81RR1936 pKa = 11.84DD1937 pKa = 3.99NTVSVGDD1944 pKa = 4.08AGTGLTRR1951 pKa = 11.84QITNVAAGTEE1961 pKa = 4.18ANDD1964 pKa = 4.01AVNKK1968 pKa = 9.86GQLDD1972 pKa = 3.96EE1973 pKa = 5.34LSDD1976 pKa = 4.3SIGDD1980 pKa = 3.62ANYY1983 pKa = 10.73YY1984 pKa = 9.66FAADD1988 pKa = 3.85GAGDD1992 pKa = 3.88GSDD1995 pKa = 3.25AAVASGVGSVAAGANSVASGTGTVAYY2021 pKa = 10.16GRR2023 pKa = 11.84GALASGEE2030 pKa = 3.96QAMAIGDD2037 pKa = 4.11GAQADD2042 pKa = 4.32YY2043 pKa = 9.88FYY2045 pKa = 11.75AMAIGAGAVSSSWASMAIGRR2065 pKa = 11.84NAIASGPRR2073 pKa = 11.84AVAIGEE2079 pKa = 4.1NAQAMDD2085 pKa = 4.11NGNVALGWASQSVGMFSAALGGSSAAIGWNTLAAGFGAWVLGNNATAVGAGAGSFYY2141 pKa = 10.66TVDD2144 pKa = 3.97EE2145 pKa = 4.24ITTVGSNARR2154 pKa = 11.84ADD2156 pKa = 3.79SFRR2159 pKa = 11.84GTVVGAGSVAAEE2171 pKa = 4.09WAPEE2175 pKa = 4.0STIVGANAFSFDD2187 pKa = 4.17SNGTAIGEE2195 pKa = 4.32SSQVSYY2201 pKa = 11.02GAANAVALGSGSHH2214 pKa = 7.05ADD2216 pKa = 3.66RR2217 pKa = 11.84ANTVSVGASGPWTDD2231 pKa = 3.43GFGVEE2236 pKa = 4.82HH2237 pKa = 7.46PALQRR2242 pKa = 11.84QITNVAAGTEE2252 pKa = 3.99ASDD2255 pKa = 4.44AVNKK2259 pKa = 10.29AQLDD2263 pKa = 3.88EE2264 pKa = 4.15VAAAGEE2270 pKa = 4.13ATGKK2274 pKa = 10.35FFF2276 pKa = 5.64
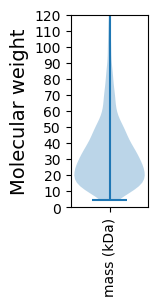
Molecular weight: 216.29 kDa
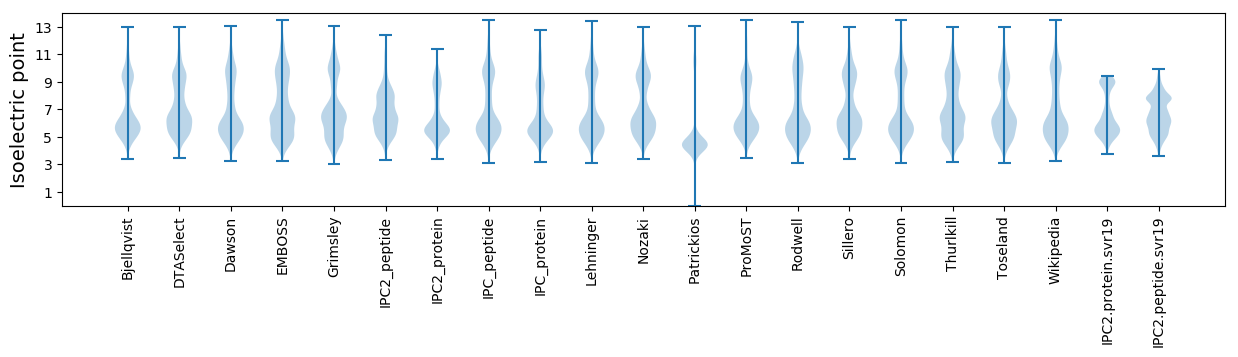
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A562LFD5|A0A562LFD5_9GAMM TetR family transcriptional regulator OS=Luteimonas cucumeris OX=985012 GN=IP90_00586 PE=4 SV=1
MM1 pKa = 7.41AVKK4 pKa = 10.23KK5 pKa = 10.04AAKK8 pKa = 9.47KK9 pKa = 10.42RR10 pKa = 11.84PAKK13 pKa = 10.19KK14 pKa = 9.78AAKK17 pKa = 8.48KK18 pKa = 7.32TAKK21 pKa = 9.03KK22 pKa = 6.66TTAKK26 pKa = 10.32KK27 pKa = 9.12ATRR30 pKa = 11.84KK31 pKa = 8.48VAKK34 pKa = 9.92KK35 pKa = 10.08AARR38 pKa = 11.84KK39 pKa = 7.18PAKK42 pKa = 9.89KK43 pKa = 9.97AVRR46 pKa = 11.84KK47 pKa = 8.88AAKK50 pKa = 10.02KK51 pKa = 9.89KK52 pKa = 8.43VAKK55 pKa = 9.97KK56 pKa = 10.19AVRR59 pKa = 11.84KK60 pKa = 8.84AVKK63 pKa = 10.2RR64 pKa = 11.84KK65 pKa = 8.03VAKK68 pKa = 10.1KK69 pKa = 9.16AAKK72 pKa = 9.71KK73 pKa = 10.15AVRR76 pKa = 11.84KK77 pKa = 8.9AAKK80 pKa = 10.02KK81 pKa = 9.89KK82 pKa = 8.43VAKK85 pKa = 10.07KK86 pKa = 10.23AVRR89 pKa = 11.84KK90 pKa = 8.72VAKK93 pKa = 10.31KK94 pKa = 10.02KK95 pKa = 8.89VAKK98 pKa = 10.27KK99 pKa = 10.06AVKK102 pKa = 9.9KK103 pKa = 10.01AAKK106 pKa = 9.87KK107 pKa = 8.93KK108 pKa = 7.13TAKK111 pKa = 8.99KK112 pKa = 7.45TAKK115 pKa = 10.09KK116 pKa = 9.81AAKK119 pKa = 9.34KK120 pKa = 10.09AKK122 pKa = 9.71KK123 pKa = 10.02KK124 pKa = 8.23PARR127 pKa = 11.84RR128 pKa = 11.84AKK130 pKa = 9.82KK131 pKa = 9.68AAPATMPAMPAPMII145 pKa = 4.36
MM1 pKa = 7.41AVKK4 pKa = 10.23KK5 pKa = 10.04AAKK8 pKa = 9.47KK9 pKa = 10.42RR10 pKa = 11.84PAKK13 pKa = 10.19KK14 pKa = 9.78AAKK17 pKa = 8.48KK18 pKa = 7.32TAKK21 pKa = 9.03KK22 pKa = 6.66TTAKK26 pKa = 10.32KK27 pKa = 9.12ATRR30 pKa = 11.84KK31 pKa = 8.48VAKK34 pKa = 9.92KK35 pKa = 10.08AARR38 pKa = 11.84KK39 pKa = 7.18PAKK42 pKa = 9.89KK43 pKa = 9.97AVRR46 pKa = 11.84KK47 pKa = 8.88AAKK50 pKa = 10.02KK51 pKa = 9.89KK52 pKa = 8.43VAKK55 pKa = 9.97KK56 pKa = 10.19AVRR59 pKa = 11.84KK60 pKa = 8.84AVKK63 pKa = 10.2RR64 pKa = 11.84KK65 pKa = 8.03VAKK68 pKa = 10.1KK69 pKa = 9.16AAKK72 pKa = 9.71KK73 pKa = 10.15AVRR76 pKa = 11.84KK77 pKa = 8.9AAKK80 pKa = 10.02KK81 pKa = 9.89KK82 pKa = 8.43VAKK85 pKa = 10.07KK86 pKa = 10.23AVRR89 pKa = 11.84KK90 pKa = 8.72VAKK93 pKa = 10.31KK94 pKa = 10.02KK95 pKa = 8.89VAKK98 pKa = 10.27KK99 pKa = 10.06AVKK102 pKa = 9.9KK103 pKa = 10.01AAKK106 pKa = 9.87KK107 pKa = 8.93KK108 pKa = 7.13TAKK111 pKa = 8.99KK112 pKa = 7.45TAKK115 pKa = 10.09KK116 pKa = 9.81AAKK119 pKa = 9.34KK120 pKa = 10.09AKK122 pKa = 9.71KK123 pKa = 10.02KK124 pKa = 8.23PARR127 pKa = 11.84RR128 pKa = 11.84AKK130 pKa = 9.82KK131 pKa = 9.68AAPATMPAMPAPMII145 pKa = 4.36
Molecular weight: 15.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1045802 |
39 |
2311 |
325.5 |
35.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.061 ± 0.079 | 0.801 ± 0.014 |
6.124 ± 0.033 | 5.305 ± 0.041 |
3.391 ± 0.027 | 8.55 ± 0.055 |
2.231 ± 0.022 | 4.324 ± 0.027 |
2.796 ± 0.038 | 10.616 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.017 | 2.554 ± 0.028 |
5.214 ± 0.037 | 3.786 ± 0.028 |
7.678 ± 0.052 | 5.315 ± 0.036 |
4.921 ± 0.035 | 7.26 ± 0.037 |
1.516 ± 0.019 | 2.349 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
