
Pseudomonas benzenivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
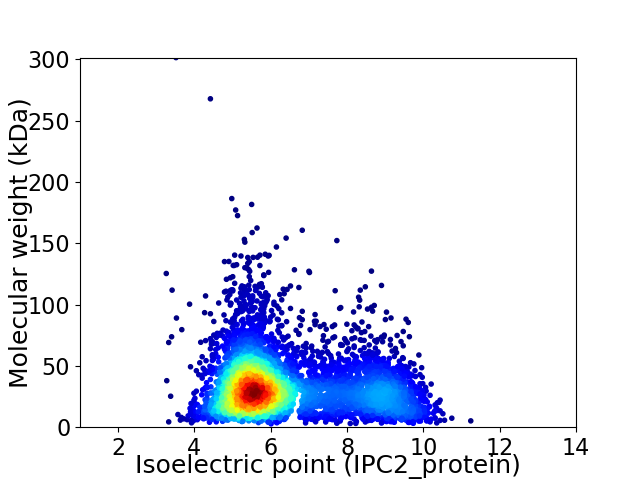
Virtual 2D-PAGE plot for 5186 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8EHF4|A0A1G8EHF4_9PSED Uncharacterized protein OS=Pseudomonas benzenivorans OX=556533 GN=SAMN05216603_11244 PE=4 SV=1
MM1 pKa = 6.91HH2 pKa = 7.08TRR4 pKa = 11.84NFFLRR9 pKa = 11.84TLPRR13 pKa = 11.84YY14 pKa = 9.76LLAAALAAGVATTGLYY30 pKa = 10.52AVDD33 pKa = 4.79GSPPGLFEE41 pKa = 6.53LDD43 pKa = 3.61GNTVDD48 pKa = 3.83VQPGGGDD55 pKa = 3.17DD56 pKa = 3.51WGSLYY61 pKa = 11.02AGDD64 pKa = 4.58QLGTPAAFTGILADD78 pKa = 4.26PAPLSIFTQGGSKK91 pKa = 10.43DD92 pKa = 3.66INDD95 pKa = 3.43VTEE98 pKa = 3.89WRR100 pKa = 11.84HH101 pKa = 4.64TDD103 pKa = 3.09GNVPDD108 pKa = 5.32KK109 pKa = 11.45DD110 pKa = 4.98DD111 pKa = 3.37ITNAYY116 pKa = 8.54AAAYY120 pKa = 6.76TVPAGGGDD128 pKa = 3.85SLNDD132 pKa = 3.98PGDD135 pKa = 3.97LIIYY139 pKa = 9.56FGLDD143 pKa = 2.68RR144 pKa = 11.84FANAGDD150 pKa = 3.59AFAGFWFFQDD160 pKa = 3.99QVGLGPNKK168 pKa = 9.95QFIGEE173 pKa = 4.21HH174 pKa = 5.38TEE176 pKa = 3.9GDD178 pKa = 3.78LLVLVEE184 pKa = 4.33YY185 pKa = 9.49PQGANAVPEE194 pKa = 4.11IKK196 pKa = 10.33VYY198 pKa = 10.37QWVASGGDD206 pKa = 3.58VAEE209 pKa = 4.88HH210 pKa = 7.04LDD212 pKa = 4.04QIFEE216 pKa = 4.34TTAQCDD222 pKa = 3.77GLGDD226 pKa = 3.91KK227 pKa = 9.65LACAITNHH235 pKa = 6.85DD236 pKa = 4.22NLSGEE241 pKa = 4.37PAWPYY246 pKa = 8.94TPKK249 pKa = 10.51SGSGLPFEE257 pKa = 4.91SFYY260 pKa = 11.4EE261 pKa = 4.17GGINVSKK268 pKa = 10.97LIGGDD273 pKa = 3.74VPCFSSFLAEE283 pKa = 4.17TRR285 pKa = 11.84SSRR288 pKa = 11.84SEE290 pKa = 3.7TAQLKK295 pKa = 10.84DD296 pKa = 3.66FVLGDD301 pKa = 4.12FDD303 pKa = 4.4LCSIAVTKK311 pKa = 10.75ACTAAVDD318 pKa = 3.88ASDD321 pKa = 4.18GGNSILVDD329 pKa = 3.94FDD331 pKa = 4.23GVVTNDD337 pKa = 3.75GGLDD341 pKa = 3.53LHH343 pKa = 7.04DD344 pKa = 3.81VTVTDD349 pKa = 5.29DD350 pKa = 3.56NGTPGDD356 pKa = 3.75TGDD359 pKa = 3.87DD360 pKa = 3.47VVVFGPADD368 pKa = 3.53LAAGEE373 pKa = 4.24SQPYY377 pKa = 8.81SGSYY381 pKa = 8.37STTAIPATDD390 pKa = 3.19TVTAEE395 pKa = 3.84GHH397 pKa = 6.2RR398 pKa = 11.84NGTSVTATADD408 pKa = 3.66ATCSPDD414 pKa = 3.27IEE416 pKa = 4.52PALTVDD422 pKa = 4.67KK423 pKa = 11.04FCTANINTGGTAIDD437 pKa = 3.5VLFNGTVTNSGNVALEE453 pKa = 4.17DD454 pKa = 3.56VTVVDD459 pKa = 4.84DD460 pKa = 5.29QGTADD465 pKa = 4.05PADD468 pKa = 4.2DD469 pKa = 3.9VTVLGPITLNVGEE482 pKa = 4.35SAPYY486 pKa = 10.04NGGFSVTGSNSSTDD500 pKa = 3.14HH501 pKa = 4.98VTATGSDD508 pKa = 3.87VLTGTPVQANAEE520 pKa = 4.31ATCQADD526 pKa = 4.01VLPAIAVDD534 pKa = 4.45KK535 pKa = 11.33VCTANINAGGTGIDD549 pKa = 3.46VLFGGTVSNTGNVALGDD566 pKa = 3.7VTVVDD571 pKa = 4.32NNGTADD577 pKa = 3.76PADD580 pKa = 4.22DD581 pKa = 3.9VTVLGPITLAPGEE594 pKa = 4.26SAPYY598 pKa = 10.12SGGFSASGSSSTDD611 pKa = 3.01MVVASGTDD619 pKa = 3.63LVTGTPVQADD629 pKa = 3.84ASASCAADD637 pKa = 3.45VLPAIDD643 pKa = 3.73VTKK646 pKa = 10.66QCTDD650 pKa = 2.93AAAFGQAILFDD661 pKa = 3.93GTVTNSGNVALLGVTVVDD679 pKa = 5.25DD680 pKa = 4.43NGTPADD686 pKa = 4.22PSDD689 pKa = 4.34DD690 pKa = 3.56VTFNLGDD697 pKa = 4.04LAPGASANYY706 pKa = 8.86NGSYY710 pKa = 9.58TPSLAGFHH718 pKa = 6.63TNTVVASASDD728 pKa = 3.59AVEE731 pKa = 4.33SGPVSATASATCEE744 pKa = 4.15VPPPPDD750 pKa = 4.17FEE752 pKa = 5.74GCTPGFWKK760 pKa = 10.5NSPGSWVGYY769 pKa = 10.41SPDD772 pKa = 3.46QLVSSVFSLPNGVLANQLGDD792 pKa = 3.86DD793 pKa = 4.16TLMEE797 pKa = 4.51ALGYY801 pKa = 9.82PGGDD805 pKa = 3.28NLVGAAQILLRR816 pKa = 11.84AAVASLLNAAHH827 pKa = 7.44PDD829 pKa = 2.9VDD831 pKa = 4.68FPRR834 pKa = 11.84TEE836 pKa = 3.92AEE838 pKa = 3.99IIADD842 pKa = 3.71VNAALATKK850 pKa = 10.43DD851 pKa = 3.17RR852 pKa = 11.84ATILALASEE861 pKa = 4.85LDD863 pKa = 3.21ADD865 pKa = 4.28NNLGCDD871 pKa = 3.74LANDD875 pKa = 3.6NSFF878 pKa = 3.47
MM1 pKa = 6.91HH2 pKa = 7.08TRR4 pKa = 11.84NFFLRR9 pKa = 11.84TLPRR13 pKa = 11.84YY14 pKa = 9.76LLAAALAAGVATTGLYY30 pKa = 10.52AVDD33 pKa = 4.79GSPPGLFEE41 pKa = 6.53LDD43 pKa = 3.61GNTVDD48 pKa = 3.83VQPGGGDD55 pKa = 3.17DD56 pKa = 3.51WGSLYY61 pKa = 11.02AGDD64 pKa = 4.58QLGTPAAFTGILADD78 pKa = 4.26PAPLSIFTQGGSKK91 pKa = 10.43DD92 pKa = 3.66INDD95 pKa = 3.43VTEE98 pKa = 3.89WRR100 pKa = 11.84HH101 pKa = 4.64TDD103 pKa = 3.09GNVPDD108 pKa = 5.32KK109 pKa = 11.45DD110 pKa = 4.98DD111 pKa = 3.37ITNAYY116 pKa = 8.54AAAYY120 pKa = 6.76TVPAGGGDD128 pKa = 3.85SLNDD132 pKa = 3.98PGDD135 pKa = 3.97LIIYY139 pKa = 9.56FGLDD143 pKa = 2.68RR144 pKa = 11.84FANAGDD150 pKa = 3.59AFAGFWFFQDD160 pKa = 3.99QVGLGPNKK168 pKa = 9.95QFIGEE173 pKa = 4.21HH174 pKa = 5.38TEE176 pKa = 3.9GDD178 pKa = 3.78LLVLVEE184 pKa = 4.33YY185 pKa = 9.49PQGANAVPEE194 pKa = 4.11IKK196 pKa = 10.33VYY198 pKa = 10.37QWVASGGDD206 pKa = 3.58VAEE209 pKa = 4.88HH210 pKa = 7.04LDD212 pKa = 4.04QIFEE216 pKa = 4.34TTAQCDD222 pKa = 3.77GLGDD226 pKa = 3.91KK227 pKa = 9.65LACAITNHH235 pKa = 6.85DD236 pKa = 4.22NLSGEE241 pKa = 4.37PAWPYY246 pKa = 8.94TPKK249 pKa = 10.51SGSGLPFEE257 pKa = 4.91SFYY260 pKa = 11.4EE261 pKa = 4.17GGINVSKK268 pKa = 10.97LIGGDD273 pKa = 3.74VPCFSSFLAEE283 pKa = 4.17TRR285 pKa = 11.84SSRR288 pKa = 11.84SEE290 pKa = 3.7TAQLKK295 pKa = 10.84DD296 pKa = 3.66FVLGDD301 pKa = 4.12FDD303 pKa = 4.4LCSIAVTKK311 pKa = 10.75ACTAAVDD318 pKa = 3.88ASDD321 pKa = 4.18GGNSILVDD329 pKa = 3.94FDD331 pKa = 4.23GVVTNDD337 pKa = 3.75GGLDD341 pKa = 3.53LHH343 pKa = 7.04DD344 pKa = 3.81VTVTDD349 pKa = 5.29DD350 pKa = 3.56NGTPGDD356 pKa = 3.75TGDD359 pKa = 3.87DD360 pKa = 3.47VVVFGPADD368 pKa = 3.53LAAGEE373 pKa = 4.24SQPYY377 pKa = 8.81SGSYY381 pKa = 8.37STTAIPATDD390 pKa = 3.19TVTAEE395 pKa = 3.84GHH397 pKa = 6.2RR398 pKa = 11.84NGTSVTATADD408 pKa = 3.66ATCSPDD414 pKa = 3.27IEE416 pKa = 4.52PALTVDD422 pKa = 4.67KK423 pKa = 11.04FCTANINTGGTAIDD437 pKa = 3.5VLFNGTVTNSGNVALEE453 pKa = 4.17DD454 pKa = 3.56VTVVDD459 pKa = 4.84DD460 pKa = 5.29QGTADD465 pKa = 4.05PADD468 pKa = 4.2DD469 pKa = 3.9VTVLGPITLNVGEE482 pKa = 4.35SAPYY486 pKa = 10.04NGGFSVTGSNSSTDD500 pKa = 3.14HH501 pKa = 4.98VTATGSDD508 pKa = 3.87VLTGTPVQANAEE520 pKa = 4.31ATCQADD526 pKa = 4.01VLPAIAVDD534 pKa = 4.45KK535 pKa = 11.33VCTANINAGGTGIDD549 pKa = 3.46VLFGGTVSNTGNVALGDD566 pKa = 3.7VTVVDD571 pKa = 4.32NNGTADD577 pKa = 3.76PADD580 pKa = 4.22DD581 pKa = 3.9VTVLGPITLAPGEE594 pKa = 4.26SAPYY598 pKa = 10.12SGGFSASGSSSTDD611 pKa = 3.01MVVASGTDD619 pKa = 3.63LVTGTPVQADD629 pKa = 3.84ASASCAADD637 pKa = 3.45VLPAIDD643 pKa = 3.73VTKK646 pKa = 10.66QCTDD650 pKa = 2.93AAAFGQAILFDD661 pKa = 3.93GTVTNSGNVALLGVTVVDD679 pKa = 5.25DD680 pKa = 4.43NGTPADD686 pKa = 4.22PSDD689 pKa = 4.34DD690 pKa = 3.56VTFNLGDD697 pKa = 4.04LAPGASANYY706 pKa = 8.86NGSYY710 pKa = 9.58TPSLAGFHH718 pKa = 6.63TNTVVASASDD728 pKa = 3.59AVEE731 pKa = 4.33SGPVSATASATCEE744 pKa = 4.15VPPPPDD750 pKa = 4.17FEE752 pKa = 5.74GCTPGFWKK760 pKa = 10.5NSPGSWVGYY769 pKa = 10.41SPDD772 pKa = 3.46QLVSSVFSLPNGVLANQLGDD792 pKa = 3.86DD793 pKa = 4.16TLMEE797 pKa = 4.51ALGYY801 pKa = 9.82PGGDD805 pKa = 3.28NLVGAAQILLRR816 pKa = 11.84AAVASLLNAAHH827 pKa = 7.44PDD829 pKa = 2.9VDD831 pKa = 4.68FPRR834 pKa = 11.84TEE836 pKa = 3.92AEE838 pKa = 3.99IIADD842 pKa = 3.71VNAALATKK850 pKa = 10.43DD851 pKa = 3.17RR852 pKa = 11.84ATILALASEE861 pKa = 4.85LDD863 pKa = 3.21ADD865 pKa = 4.28NNLGCDD871 pKa = 3.74LANDD875 pKa = 3.6NSFF878 pKa = 3.47
Molecular weight: 89.06 kDa
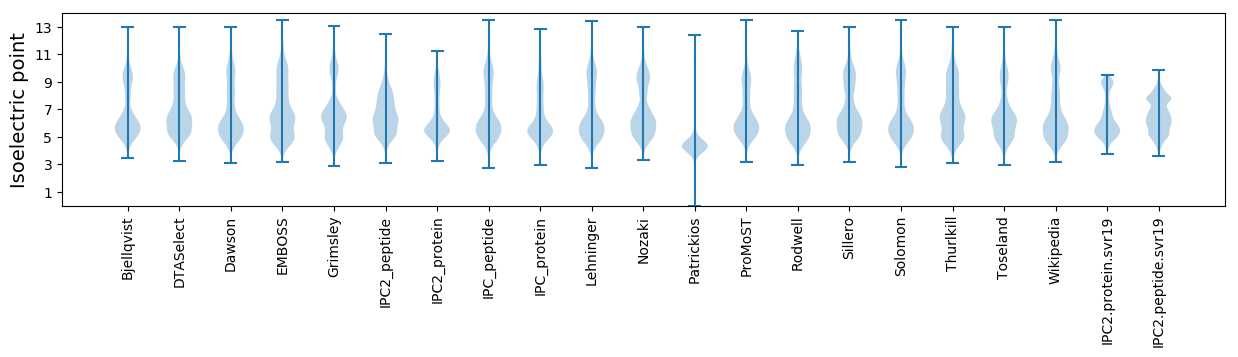
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8BVY8|A0A1G8BVY8_9PSED UPF0276 protein SAMN05216603_108176 OS=Pseudomonas benzenivorans OX=556533 GN=SAMN05216603_108176 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1669955 |
26 |
3143 |
322.0 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.916 ± 0.045 | 1.056 ± 0.012 |
5.144 ± 0.025 | 5.927 ± 0.036 |
3.577 ± 0.02 | 8.137 ± 0.036 |
2.251 ± 0.019 | 4.486 ± 0.029 |
3.077 ± 0.031 | 12.54 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.128 ± 0.016 | 2.661 ± 0.021 |
4.941 ± 0.026 | 4.745 ± 0.03 |
6.904 ± 0.037 | 5.575 ± 0.028 |
4.239 ± 0.028 | 6.763 ± 0.028 |
1.444 ± 0.017 | 2.488 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
