
Wenling hepe-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
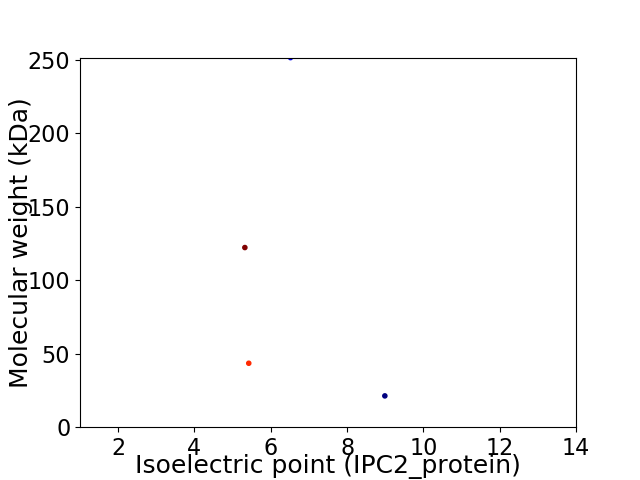
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KKG8|A0A1L3KKG8_9VIRU Putative glycoprotein OS=Wenling hepe-like virus 2 OX=1923494 PE=4 SV=1
MM1 pKa = 7.39ILSLKK6 pKa = 9.03PLKK9 pKa = 9.48STRR12 pKa = 11.84ICQKK16 pKa = 9.02STLLEE21 pKa = 3.93SVKK24 pKa = 10.61FIEE27 pKa = 4.55SAEE30 pKa = 3.94FVINNFDD37 pKa = 3.57PKK39 pKa = 10.66MNIPAQWVNAANFPRR54 pKa = 11.84KK55 pKa = 9.67GGSAAPYY62 pKa = 10.38VNIANLPDD70 pKa = 5.05DD71 pKa = 4.4LTGQQPSLGDD81 pKa = 4.0ANNLAAMVAILQAARR96 pKa = 11.84YY97 pKa = 8.42KK98 pKa = 10.78ADD100 pKa = 3.63EE101 pKa = 4.58AFSDD105 pKa = 3.89ATAQLQLDD113 pKa = 4.18TVTSDD118 pKa = 3.2EE119 pKa = 4.52LSFVPLVSNVHH130 pKa = 6.14EE131 pKa = 4.22LTPSNFTHH139 pKa = 5.98SWAVSFPPGIKK150 pKa = 9.11QDD152 pKa = 3.68LYY154 pKa = 9.47FTRR157 pKa = 11.84GPTPTGPGQYY167 pKa = 9.98QFYY170 pKa = 10.64NEE172 pKa = 4.36TFVEE176 pKa = 4.69SDD178 pKa = 3.99SGWITQKK185 pKa = 9.74TGSQDD190 pKa = 3.93AIPALPEE197 pKa = 4.21SIQIRR202 pKa = 11.84RR203 pKa = 11.84GGRR206 pKa = 11.84NTFFKK211 pKa = 10.64IKK213 pKa = 9.89GYY215 pKa = 10.85NKK217 pKa = 9.1IRR219 pKa = 11.84LVASTDD225 pKa = 3.17AQQLVTVVCDD235 pKa = 3.77PYY237 pKa = 11.28GVRR240 pKa = 11.84IYY242 pKa = 11.1AGLPFKK248 pKa = 10.59EE249 pKa = 4.41PQLLAQTRR257 pKa = 11.84TVAYY261 pKa = 9.52LRR263 pKa = 11.84NSTVVVGVPGVSEE276 pKa = 3.66ITMIAGYY283 pKa = 8.94VHH285 pKa = 7.12AWADD289 pKa = 3.75TNFSEE294 pKa = 5.61FGFHH298 pKa = 7.62GDD300 pKa = 3.2VEE302 pKa = 4.46KK303 pKa = 10.8CARR306 pKa = 11.84LSALEE311 pKa = 4.09LFYY314 pKa = 10.47TAVRR318 pKa = 11.84AAVGQSVIKK327 pKa = 10.45AALNASSLTQKK338 pKa = 10.29VSALTPALNARR349 pKa = 11.84MVTSLAKK356 pKa = 9.88NAIMGISEE364 pKa = 4.29SFEE367 pKa = 4.27AEE369 pKa = 3.77APFASGQTRR378 pKa = 11.84DD379 pKa = 3.62NAIHH383 pKa = 6.76GVTLVPFGGDD393 pKa = 3.17DD394 pKa = 3.93SDD396 pKa = 4.01PKK398 pKa = 11.01EE399 pKa = 4.03LDD401 pKa = 3.84FILRR405 pKa = 11.84GPIQPFSPWYY415 pKa = 9.21EE416 pKa = 3.98ATSSSGVKK424 pKa = 9.74FRR426 pKa = 11.84TGMVVDD432 pKa = 5.76EE433 pKa = 4.37FAGWSTGTGVGQVTITANHH452 pKa = 6.25EE453 pKa = 4.05SGYY456 pKa = 11.2VNFTPEE462 pKa = 4.72DD463 pKa = 3.3PWNNNLYY470 pKa = 8.97VSKK473 pKa = 10.31RR474 pKa = 11.84VEE476 pKa = 4.0VFLEE480 pKa = 4.18SPTVAGAGFEE490 pKa = 4.3ASGTATYY497 pKa = 10.11KK498 pKa = 10.85GKK500 pKa = 8.97TYY502 pKa = 10.87QLVFDD507 pKa = 4.12VTEE510 pKa = 4.24EE511 pKa = 4.47DD512 pKa = 4.13DD513 pKa = 5.63LITAFASVTLEE524 pKa = 3.9QTHH527 pKa = 6.05FEE529 pKa = 4.06PWVLQNFTTYY539 pKa = 9.73MGTGCSIKK547 pKa = 10.6NQEE550 pKa = 3.42QCYY553 pKa = 10.0IGEE556 pKa = 4.26TEE558 pKa = 4.87HH559 pKa = 8.05IYY561 pKa = 10.77GYY563 pKa = 10.58DD564 pKa = 3.65GPDD567 pKa = 3.39PLMVKK572 pKa = 10.47ALVDD576 pKa = 3.8YY577 pKa = 10.65EE578 pKa = 4.47IGTSDD583 pKa = 3.11STTYY587 pKa = 11.06LHH589 pKa = 6.65RR590 pKa = 11.84CKK592 pKa = 10.45HH593 pKa = 5.74RR594 pKa = 11.84LSPPWNTNLSLEE606 pKa = 4.43TKK608 pKa = 9.98PVINVKK614 pKa = 9.99VYY616 pKa = 10.6SVPPVTAAPVVVDD629 pKa = 3.61TTEE632 pKa = 3.94TIMFKK637 pKa = 10.72EE638 pKa = 4.14LVLTSLNSAVNSVDD652 pKa = 3.37IRR654 pKa = 11.84QQDD657 pKa = 3.13RR658 pKa = 11.84RR659 pKa = 11.84IEE661 pKa = 4.32EE662 pKa = 3.79IEE664 pKa = 3.95RR665 pKa = 11.84RR666 pKa = 11.84TRR668 pKa = 11.84PTVAGFISTAAFTAASFLNPGVGLTAALLVGTVGEE703 pKa = 4.2AVEE706 pKa = 4.68SFQHH710 pKa = 6.87GYY712 pKa = 8.46TIQSIAEE719 pKa = 4.14MVTAGLVHH727 pKa = 6.04TAQRR731 pKa = 11.84RR732 pKa = 11.84VHH734 pKa = 6.56GKK736 pKa = 10.44DD737 pKa = 3.25EE738 pKa = 4.17VDD740 pKa = 3.5EE741 pKa = 5.19SDD743 pKa = 4.21DD744 pKa = 3.82FLVSVLNILLPSKK757 pKa = 10.66ASSVKK762 pKa = 10.22LAEE765 pKa = 4.37HH766 pKa = 6.06RR767 pKa = 11.84TKK769 pKa = 10.89VMKK772 pKa = 10.4SIEE775 pKa = 4.14RR776 pKa = 11.84GEE778 pKa = 4.25PLTVPTAQGFPVGHH792 pKa = 7.06KK793 pKa = 10.21SLSLPFVSLNNKK805 pKa = 9.38QITPIKK811 pKa = 10.25VPLPSEE817 pKa = 4.38VPPKK821 pKa = 9.9FQPYY825 pKa = 10.36ANKK828 pKa = 10.57LEE830 pKa = 4.3NKK832 pKa = 9.51GMYY835 pKa = 9.18PIHH838 pKa = 6.83QSVTANSVMEE848 pKa = 4.49VGDD851 pKa = 3.57QTVGFQISLGVSDD864 pKa = 4.61GVQRR868 pKa = 11.84GDD870 pKa = 3.17GVFPSRR876 pKa = 11.84GGVKK880 pKa = 10.1GVPSEE885 pKa = 4.24PGCSFFLHH893 pKa = 7.38DD894 pKa = 5.12GGISNNVLSIRR905 pKa = 11.84EE906 pKa = 4.03FTSDD910 pKa = 3.04SNIIFHH916 pKa = 6.3EE917 pKa = 4.03NRR919 pKa = 11.84AFMAQLGFRR928 pKa = 11.84KK929 pKa = 9.95SYY931 pKa = 10.48IDD933 pKa = 3.38SLSPEE938 pKa = 4.12RR939 pKa = 11.84MDD941 pKa = 4.84QILKK945 pKa = 10.25IPTVSQMALKK955 pKa = 9.31TVGSEE960 pKa = 3.93RR961 pKa = 11.84CSDD964 pKa = 4.17FSLSLHH970 pKa = 6.25SMASIVQSHH979 pKa = 6.14TGAAGRR985 pKa = 11.84EE986 pKa = 3.76MWGYY990 pKa = 10.8NVLNHH995 pKa = 6.11NCRR998 pKa = 11.84HH999 pKa = 5.27YY1000 pKa = 11.16ARR1002 pKa = 11.84EE1003 pKa = 3.95AFEE1006 pKa = 4.68TIMTGTPQGQVVPWFHH1022 pKa = 7.26RR1023 pKa = 11.84YY1024 pKa = 8.75IKK1026 pKa = 10.33EE1027 pKa = 3.82RR1028 pKa = 11.84ASNNMSRR1035 pKa = 11.84EE1036 pKa = 3.96EE1037 pKa = 3.94FVDD1040 pKa = 3.92VVDD1043 pKa = 5.7NIYY1046 pKa = 10.29QHH1048 pKa = 7.06IYY1050 pKa = 8.34HH1051 pKa = 6.74TLDD1054 pKa = 5.03DD1055 pKa = 4.66IDD1057 pKa = 4.1NSVYY1061 pKa = 10.44DD1062 pKa = 4.14IKK1064 pKa = 11.21HH1065 pKa = 6.84LIATMYY1071 pKa = 11.09GEE1073 pKa = 4.22TLTDD1077 pKa = 3.55EE1078 pKa = 4.46EE1079 pKa = 5.05EE1080 pKa = 4.4EE1081 pKa = 4.27IVDD1084 pKa = 4.09NKK1086 pKa = 10.17YY1087 pKa = 11.14LKK1089 pKa = 10.48DD1090 pKa = 3.65DD1091 pKa = 3.66SKK1093 pKa = 11.48EE1094 pKa = 4.0PRR1096 pKa = 11.84EE1097 pKa = 3.86HH1098 pKa = 6.18TFRR1101 pKa = 11.84NGRR1104 pKa = 11.84PGKK1107 pKa = 10.15LLSFNN1112 pKa = 4.03
MM1 pKa = 7.39ILSLKK6 pKa = 9.03PLKK9 pKa = 9.48STRR12 pKa = 11.84ICQKK16 pKa = 9.02STLLEE21 pKa = 3.93SVKK24 pKa = 10.61FIEE27 pKa = 4.55SAEE30 pKa = 3.94FVINNFDD37 pKa = 3.57PKK39 pKa = 10.66MNIPAQWVNAANFPRR54 pKa = 11.84KK55 pKa = 9.67GGSAAPYY62 pKa = 10.38VNIANLPDD70 pKa = 5.05DD71 pKa = 4.4LTGQQPSLGDD81 pKa = 4.0ANNLAAMVAILQAARR96 pKa = 11.84YY97 pKa = 8.42KK98 pKa = 10.78ADD100 pKa = 3.63EE101 pKa = 4.58AFSDD105 pKa = 3.89ATAQLQLDD113 pKa = 4.18TVTSDD118 pKa = 3.2EE119 pKa = 4.52LSFVPLVSNVHH130 pKa = 6.14EE131 pKa = 4.22LTPSNFTHH139 pKa = 5.98SWAVSFPPGIKK150 pKa = 9.11QDD152 pKa = 3.68LYY154 pKa = 9.47FTRR157 pKa = 11.84GPTPTGPGQYY167 pKa = 9.98QFYY170 pKa = 10.64NEE172 pKa = 4.36TFVEE176 pKa = 4.69SDD178 pKa = 3.99SGWITQKK185 pKa = 9.74TGSQDD190 pKa = 3.93AIPALPEE197 pKa = 4.21SIQIRR202 pKa = 11.84RR203 pKa = 11.84GGRR206 pKa = 11.84NTFFKK211 pKa = 10.64IKK213 pKa = 9.89GYY215 pKa = 10.85NKK217 pKa = 9.1IRR219 pKa = 11.84LVASTDD225 pKa = 3.17AQQLVTVVCDD235 pKa = 3.77PYY237 pKa = 11.28GVRR240 pKa = 11.84IYY242 pKa = 11.1AGLPFKK248 pKa = 10.59EE249 pKa = 4.41PQLLAQTRR257 pKa = 11.84TVAYY261 pKa = 9.52LRR263 pKa = 11.84NSTVVVGVPGVSEE276 pKa = 3.66ITMIAGYY283 pKa = 8.94VHH285 pKa = 7.12AWADD289 pKa = 3.75TNFSEE294 pKa = 5.61FGFHH298 pKa = 7.62GDD300 pKa = 3.2VEE302 pKa = 4.46KK303 pKa = 10.8CARR306 pKa = 11.84LSALEE311 pKa = 4.09LFYY314 pKa = 10.47TAVRR318 pKa = 11.84AAVGQSVIKK327 pKa = 10.45AALNASSLTQKK338 pKa = 10.29VSALTPALNARR349 pKa = 11.84MVTSLAKK356 pKa = 9.88NAIMGISEE364 pKa = 4.29SFEE367 pKa = 4.27AEE369 pKa = 3.77APFASGQTRR378 pKa = 11.84DD379 pKa = 3.62NAIHH383 pKa = 6.76GVTLVPFGGDD393 pKa = 3.17DD394 pKa = 3.93SDD396 pKa = 4.01PKK398 pKa = 11.01EE399 pKa = 4.03LDD401 pKa = 3.84FILRR405 pKa = 11.84GPIQPFSPWYY415 pKa = 9.21EE416 pKa = 3.98ATSSSGVKK424 pKa = 9.74FRR426 pKa = 11.84TGMVVDD432 pKa = 5.76EE433 pKa = 4.37FAGWSTGTGVGQVTITANHH452 pKa = 6.25EE453 pKa = 4.05SGYY456 pKa = 11.2VNFTPEE462 pKa = 4.72DD463 pKa = 3.3PWNNNLYY470 pKa = 8.97VSKK473 pKa = 10.31RR474 pKa = 11.84VEE476 pKa = 4.0VFLEE480 pKa = 4.18SPTVAGAGFEE490 pKa = 4.3ASGTATYY497 pKa = 10.11KK498 pKa = 10.85GKK500 pKa = 8.97TYY502 pKa = 10.87QLVFDD507 pKa = 4.12VTEE510 pKa = 4.24EE511 pKa = 4.47DD512 pKa = 4.13DD513 pKa = 5.63LITAFASVTLEE524 pKa = 3.9QTHH527 pKa = 6.05FEE529 pKa = 4.06PWVLQNFTTYY539 pKa = 9.73MGTGCSIKK547 pKa = 10.6NQEE550 pKa = 3.42QCYY553 pKa = 10.0IGEE556 pKa = 4.26TEE558 pKa = 4.87HH559 pKa = 8.05IYY561 pKa = 10.77GYY563 pKa = 10.58DD564 pKa = 3.65GPDD567 pKa = 3.39PLMVKK572 pKa = 10.47ALVDD576 pKa = 3.8YY577 pKa = 10.65EE578 pKa = 4.47IGTSDD583 pKa = 3.11STTYY587 pKa = 11.06LHH589 pKa = 6.65RR590 pKa = 11.84CKK592 pKa = 10.45HH593 pKa = 5.74RR594 pKa = 11.84LSPPWNTNLSLEE606 pKa = 4.43TKK608 pKa = 9.98PVINVKK614 pKa = 9.99VYY616 pKa = 10.6SVPPVTAAPVVVDD629 pKa = 3.61TTEE632 pKa = 3.94TIMFKK637 pKa = 10.72EE638 pKa = 4.14LVLTSLNSAVNSVDD652 pKa = 3.37IRR654 pKa = 11.84QQDD657 pKa = 3.13RR658 pKa = 11.84RR659 pKa = 11.84IEE661 pKa = 4.32EE662 pKa = 3.79IEE664 pKa = 3.95RR665 pKa = 11.84RR666 pKa = 11.84TRR668 pKa = 11.84PTVAGFISTAAFTAASFLNPGVGLTAALLVGTVGEE703 pKa = 4.2AVEE706 pKa = 4.68SFQHH710 pKa = 6.87GYY712 pKa = 8.46TIQSIAEE719 pKa = 4.14MVTAGLVHH727 pKa = 6.04TAQRR731 pKa = 11.84RR732 pKa = 11.84VHH734 pKa = 6.56GKK736 pKa = 10.44DD737 pKa = 3.25EE738 pKa = 4.17VDD740 pKa = 3.5EE741 pKa = 5.19SDD743 pKa = 4.21DD744 pKa = 3.82FLVSVLNILLPSKK757 pKa = 10.66ASSVKK762 pKa = 10.22LAEE765 pKa = 4.37HH766 pKa = 6.06RR767 pKa = 11.84TKK769 pKa = 10.89VMKK772 pKa = 10.4SIEE775 pKa = 4.14RR776 pKa = 11.84GEE778 pKa = 4.25PLTVPTAQGFPVGHH792 pKa = 7.06KK793 pKa = 10.21SLSLPFVSLNNKK805 pKa = 9.38QITPIKK811 pKa = 10.25VPLPSEE817 pKa = 4.38VPPKK821 pKa = 9.9FQPYY825 pKa = 10.36ANKK828 pKa = 10.57LEE830 pKa = 4.3NKK832 pKa = 9.51GMYY835 pKa = 9.18PIHH838 pKa = 6.83QSVTANSVMEE848 pKa = 4.49VGDD851 pKa = 3.57QTVGFQISLGVSDD864 pKa = 4.61GVQRR868 pKa = 11.84GDD870 pKa = 3.17GVFPSRR876 pKa = 11.84GGVKK880 pKa = 10.1GVPSEE885 pKa = 4.24PGCSFFLHH893 pKa = 7.38DD894 pKa = 5.12GGISNNVLSIRR905 pKa = 11.84EE906 pKa = 4.03FTSDD910 pKa = 3.04SNIIFHH916 pKa = 6.3EE917 pKa = 4.03NRR919 pKa = 11.84AFMAQLGFRR928 pKa = 11.84KK929 pKa = 9.95SYY931 pKa = 10.48IDD933 pKa = 3.38SLSPEE938 pKa = 4.12RR939 pKa = 11.84MDD941 pKa = 4.84QILKK945 pKa = 10.25IPTVSQMALKK955 pKa = 9.31TVGSEE960 pKa = 3.93RR961 pKa = 11.84CSDD964 pKa = 4.17FSLSLHH970 pKa = 6.25SMASIVQSHH979 pKa = 6.14TGAAGRR985 pKa = 11.84EE986 pKa = 3.76MWGYY990 pKa = 10.8NVLNHH995 pKa = 6.11NCRR998 pKa = 11.84HH999 pKa = 5.27YY1000 pKa = 11.16ARR1002 pKa = 11.84EE1003 pKa = 3.95AFEE1006 pKa = 4.68TIMTGTPQGQVVPWFHH1022 pKa = 7.26RR1023 pKa = 11.84YY1024 pKa = 8.75IKK1026 pKa = 10.33EE1027 pKa = 3.82RR1028 pKa = 11.84ASNNMSRR1035 pKa = 11.84EE1036 pKa = 3.96EE1037 pKa = 3.94FVDD1040 pKa = 3.92VVDD1043 pKa = 5.7NIYY1046 pKa = 10.29QHH1048 pKa = 7.06IYY1050 pKa = 8.34HH1051 pKa = 6.74TLDD1054 pKa = 5.03DD1055 pKa = 4.66IDD1057 pKa = 4.1NSVYY1061 pKa = 10.44DD1062 pKa = 4.14IKK1064 pKa = 11.21HH1065 pKa = 6.84LIATMYY1071 pKa = 11.09GEE1073 pKa = 4.22TLTDD1077 pKa = 3.55EE1078 pKa = 4.46EE1079 pKa = 5.05EE1080 pKa = 4.4EE1081 pKa = 4.27IVDD1084 pKa = 4.09NKK1086 pKa = 10.17YY1087 pKa = 11.14LKK1089 pKa = 10.48DD1090 pKa = 3.65DD1091 pKa = 3.66SKK1093 pKa = 11.48EE1094 pKa = 4.0PRR1096 pKa = 11.84EE1097 pKa = 3.86HH1098 pKa = 6.18TFRR1101 pKa = 11.84NGRR1104 pKa = 11.84PGKK1107 pKa = 10.15LLSFNN1112 pKa = 4.03
Molecular weight: 122.19 kDa
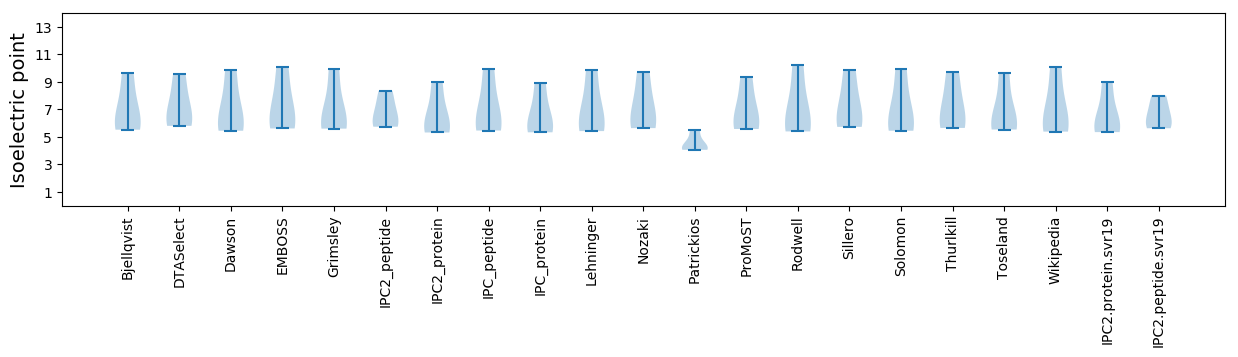
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKG8|A0A1L3KKG8_9VIRU Putative glycoprotein OS=Wenling hepe-like virus 2 OX=1923494 PE=4 SV=1
MM1 pKa = 7.85ADD3 pKa = 3.36STQEE7 pKa = 3.92STSMAKK13 pKa = 10.45YY14 pKa = 9.74KK15 pKa = 10.69EE16 pKa = 4.39SVSKK20 pKa = 10.76IMNALLHH27 pKa = 6.43FGAILHH33 pKa = 6.76LDD35 pKa = 2.82KK36 pKa = 11.43SMCVFRR42 pKa = 11.84LHH44 pKa = 5.91QNPVYY49 pKa = 10.5RR50 pKa = 11.84IVLKK54 pKa = 9.82ATYY57 pKa = 10.12KK58 pKa = 10.65DD59 pKa = 3.22ASGDD63 pKa = 3.59KK64 pKa = 10.7KK65 pKa = 10.89EE66 pKa = 4.7LEE68 pKa = 4.33TVSWGTKK75 pKa = 8.68SQANHH80 pKa = 5.48YY81 pKa = 10.0ARR83 pKa = 11.84SKK85 pKa = 10.47LLDD88 pKa = 4.16IIFHH92 pKa = 5.85EE93 pKa = 4.25TDD95 pKa = 3.25FVKK98 pKa = 10.59RR99 pKa = 11.84VDD101 pKa = 4.64LEE103 pKa = 4.46AFDD106 pKa = 5.64LLISTKK112 pKa = 10.26RR113 pKa = 11.84RR114 pKa = 11.84LTKK117 pKa = 9.26FASRR121 pKa = 11.84EE122 pKa = 3.85RR123 pKa = 11.84CTCFSLGTRR132 pKa = 11.84RR133 pKa = 11.84HH134 pKa = 6.38HH135 pKa = 5.28YY136 pKa = 8.57TEE138 pKa = 3.74KK139 pKa = 10.27CKK141 pKa = 10.1TFRR144 pKa = 11.84RR145 pKa = 11.84HH146 pKa = 5.1TNFEE150 pKa = 3.93VPLPRR155 pKa = 11.84RR156 pKa = 11.84QDD158 pKa = 3.43YY159 pKa = 9.56PFLPEE164 pKa = 3.57TRR166 pKa = 11.84YY167 pKa = 9.15FAAVAEE173 pKa = 4.69RR174 pKa = 11.84YY175 pKa = 8.82GHH177 pKa = 5.52NHH179 pKa = 4.92QFF181 pKa = 3.33
MM1 pKa = 7.85ADD3 pKa = 3.36STQEE7 pKa = 3.92STSMAKK13 pKa = 10.45YY14 pKa = 9.74KK15 pKa = 10.69EE16 pKa = 4.39SVSKK20 pKa = 10.76IMNALLHH27 pKa = 6.43FGAILHH33 pKa = 6.76LDD35 pKa = 2.82KK36 pKa = 11.43SMCVFRR42 pKa = 11.84LHH44 pKa = 5.91QNPVYY49 pKa = 10.5RR50 pKa = 11.84IVLKK54 pKa = 9.82ATYY57 pKa = 10.12KK58 pKa = 10.65DD59 pKa = 3.22ASGDD63 pKa = 3.59KK64 pKa = 10.7KK65 pKa = 10.89EE66 pKa = 4.7LEE68 pKa = 4.33TVSWGTKK75 pKa = 8.68SQANHH80 pKa = 5.48YY81 pKa = 10.0ARR83 pKa = 11.84SKK85 pKa = 10.47LLDD88 pKa = 4.16IIFHH92 pKa = 5.85EE93 pKa = 4.25TDD95 pKa = 3.25FVKK98 pKa = 10.59RR99 pKa = 11.84VDD101 pKa = 4.64LEE103 pKa = 4.46AFDD106 pKa = 5.64LLISTKK112 pKa = 10.26RR113 pKa = 11.84RR114 pKa = 11.84LTKK117 pKa = 9.26FASRR121 pKa = 11.84EE122 pKa = 3.85RR123 pKa = 11.84CTCFSLGTRR132 pKa = 11.84RR133 pKa = 11.84HH134 pKa = 6.38HH135 pKa = 5.28YY136 pKa = 8.57TEE138 pKa = 3.74KK139 pKa = 10.27CKK141 pKa = 10.1TFRR144 pKa = 11.84RR145 pKa = 11.84HH146 pKa = 5.1TNFEE150 pKa = 3.93VPLPRR155 pKa = 11.84RR156 pKa = 11.84QDD158 pKa = 3.43YY159 pKa = 9.56PFLPEE164 pKa = 3.57TRR166 pKa = 11.84YY167 pKa = 9.15FAAVAEE173 pKa = 4.69RR174 pKa = 11.84YY175 pKa = 8.82GHH177 pKa = 5.52NHH179 pKa = 4.92QFF181 pKa = 3.33
Molecular weight: 21.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3908 |
181 |
2222 |
977.0 |
109.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.883 ± 0.405 | 1.535 ± 0.316 |
5.962 ± 0.486 | 5.553 ± 0.358 |
4.836 ± 0.295 | 5.604 ± 0.583 |
2.636 ± 0.243 | 5.143 ± 0.177 |
5.732 ± 0.587 | 7.088 ± 0.375 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.277 ± 0.152 | 5.425 ± 0.434 |
5.45 ± 0.314 | 3.838 ± 0.166 |
5.118 ± 0.443 | 7.267 ± 0.428 |
7.702 ± 0.788 | 7.293 ± 0.638 |
0.896 ± 0.06 | 3.762 ± 0.526 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
