
Variovorax sp. PAMC 28711
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Variovorax; unclassified Variovorax
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
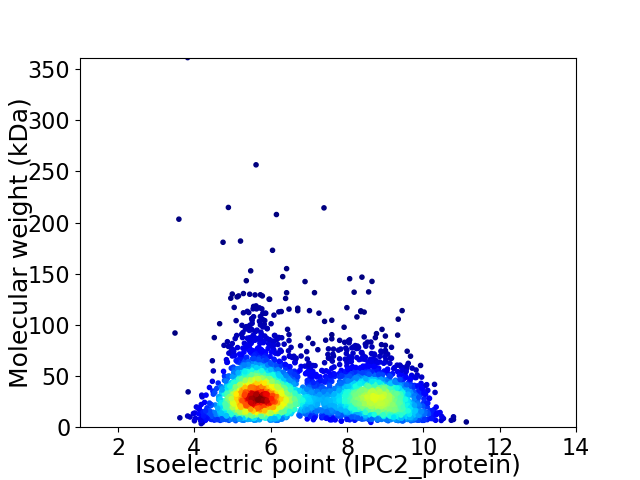
Virtual 2D-PAGE plot for 3960 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126ZF94|A0A126ZF94_9BURK Uncharacterized protein OS=Variovorax sp. PAMC 28711 OX=1795631 GN=AX767_12550 PE=4 SV=1
MM1 pKa = 7.63SYY3 pKa = 10.9YY4 pKa = 10.59VIPAGQAGQLGALGSAHH21 pKa = 7.5LDD23 pKa = 3.6GNLGAFDD30 pKa = 4.5FKK32 pKa = 11.72YY33 pKa = 10.71EE34 pKa = 4.44VIDD37 pKa = 3.45SAYY40 pKa = 9.88GLTPAGSIDD49 pKa = 3.74TVLKK53 pKa = 10.96DD54 pKa = 3.46GTLTLSATPVTDD66 pKa = 3.81AVNLSITGISGAASISDD83 pKa = 3.52EE84 pKa = 4.41VQGDD88 pKa = 3.72DD89 pKa = 4.86ANPDD93 pKa = 3.72TAVVDD98 pKa = 3.87AAGTTVTVNLNIASVDD114 pKa = 3.49YY115 pKa = 11.01DD116 pKa = 3.32GSEE119 pKa = 3.42RR120 pKa = 11.84VMRR123 pKa = 11.84VLIEE127 pKa = 3.96GVPDD131 pKa = 3.96GVTVNAPVNGQAQQVGIGTWVLVFAANALPINAAGGITLGVQFLTGVDD179 pKa = 3.72VAQATHH185 pKa = 7.96PITMTVQTQDD195 pKa = 2.76RR196 pKa = 11.84GAVTSGQAQDD206 pKa = 3.34SVTWNLVTTYY216 pKa = 10.82DD217 pKa = 3.62GAGEE221 pKa = 4.3SLPPTIDD228 pKa = 2.72QWVYY232 pKa = 10.96NGADD236 pKa = 3.51VNEE239 pKa = 4.94DD240 pKa = 3.58LTFSLSDD247 pKa = 3.44VVDD250 pKa = 4.06AQVTVVDD257 pKa = 3.89VSVPNIFTVSLTDD270 pKa = 3.87LPEE273 pKa = 4.39GTQVQGMVLTSVNGVPTWTATVVVPAGANDD303 pKa = 3.8AAVEE307 pKa = 3.9LALEE311 pKa = 4.37GLLNGIQITPPPNSNEE327 pKa = 3.86NNAEE331 pKa = 3.87DD332 pKa = 4.07AFNFNATLTASAQGSFSVSQTIPDD356 pKa = 3.84TVMVIPVAPVTDD368 pKa = 3.53AAVITVSAVDD378 pKa = 3.39AGEE381 pKa = 4.18NPVDD385 pKa = 3.68GSVSVTIDD393 pKa = 3.22VRR395 pKa = 11.84SNPVDD400 pKa = 3.28GSNGVIVGGLLYY412 pKa = 10.65VQVSATGGGNAGGTLTYY429 pKa = 10.27QGNPAALTPVTNPPGVPAGTYY450 pKa = 9.13YY451 pKa = 10.98LVPVGPSGDD460 pKa = 3.91SVNLVYY466 pKa = 10.29TPPSGTSPGNVIFTALANTQEE487 pKa = 3.97TGAAVVTGSASDD499 pKa = 3.39TATISAVNNGVTVTGFSTPVSAPEE523 pKa = 3.92TDD525 pKa = 3.49GSGLGTAIALPGLMVNLIDD544 pKa = 4.33SDD546 pKa = 3.76GSEE549 pKa = 4.41SIKK552 pKa = 10.83SVTLAGVPVGFLVYY566 pKa = 10.56VGGQLATNAGGNGTSNTWVISNSGTFGEE594 pKa = 4.66VKK596 pKa = 10.05IAPPAHH602 pKa = 6.73WSGEE606 pKa = 4.12LNGLKK611 pKa = 10.36LVVEE615 pKa = 4.49SGEE618 pKa = 4.04NLLAKK623 pKa = 10.35SLEE626 pKa = 4.16EE627 pKa = 3.8FLLPSVTVTPVASGLTLDD645 pKa = 3.51TTQAVGRR652 pKa = 11.84EE653 pKa = 4.05GGIIGLNLNASMTDD667 pKa = 3.38PVAATSTLPDD677 pKa = 3.43SSTEE681 pKa = 4.06TVTVQFKK688 pKa = 11.25GLGEE692 pKa = 3.93YY693 pKa = 10.49AAFYY697 pKa = 10.95VDD699 pKa = 4.04STLLTDD705 pKa = 4.16SGTSGHH711 pKa = 6.45TIGYY715 pKa = 9.78NDD717 pKa = 3.98LTDD720 pKa = 3.65TYY722 pKa = 9.74TITGLTQVEE731 pKa = 4.06MDD733 pKa = 3.3QLGFKK738 pKa = 9.69QAASALVDD746 pKa = 3.37QDD748 pKa = 3.7GAAGTQIEE756 pKa = 4.55VTAWTVEE763 pKa = 4.26SGGGPISAVEE773 pKa = 4.03TADD776 pKa = 2.83ITVNMTVVQPTSGADD791 pKa = 3.24SFIWGGEE798 pKa = 3.67GKK800 pKa = 10.55AIINGAAGDD809 pKa = 3.85DD810 pKa = 3.94TVALRR815 pKa = 11.84LGEE818 pKa = 4.38DD819 pKa = 3.32VSGANLAMGLRR830 pKa = 11.84NIEE833 pKa = 4.08VLDD836 pKa = 4.61LSAQGEE842 pKa = 4.31NAVTNLTATDD852 pKa = 3.76VLSLTGSNHH861 pKa = 6.24ILTINGTGDD870 pKa = 3.87DD871 pKa = 3.99SVQLGGGASAWTAGASSGGYY891 pKa = 8.01TDD893 pKa = 3.69YY894 pKa = 11.35TSGAGASLVTVRR906 pKa = 11.84IDD908 pKa = 3.98DD909 pKa = 5.61DD910 pKa = 3.57ITHH913 pKa = 6.65SYY915 pKa = 8.28VV916 pKa = 2.94
MM1 pKa = 7.63SYY3 pKa = 10.9YY4 pKa = 10.59VIPAGQAGQLGALGSAHH21 pKa = 7.5LDD23 pKa = 3.6GNLGAFDD30 pKa = 4.5FKK32 pKa = 11.72YY33 pKa = 10.71EE34 pKa = 4.44VIDD37 pKa = 3.45SAYY40 pKa = 9.88GLTPAGSIDD49 pKa = 3.74TVLKK53 pKa = 10.96DD54 pKa = 3.46GTLTLSATPVTDD66 pKa = 3.81AVNLSITGISGAASISDD83 pKa = 3.52EE84 pKa = 4.41VQGDD88 pKa = 3.72DD89 pKa = 4.86ANPDD93 pKa = 3.72TAVVDD98 pKa = 3.87AAGTTVTVNLNIASVDD114 pKa = 3.49YY115 pKa = 11.01DD116 pKa = 3.32GSEE119 pKa = 3.42RR120 pKa = 11.84VMRR123 pKa = 11.84VLIEE127 pKa = 3.96GVPDD131 pKa = 3.96GVTVNAPVNGQAQQVGIGTWVLVFAANALPINAAGGITLGVQFLTGVDD179 pKa = 3.72VAQATHH185 pKa = 7.96PITMTVQTQDD195 pKa = 2.76RR196 pKa = 11.84GAVTSGQAQDD206 pKa = 3.34SVTWNLVTTYY216 pKa = 10.82DD217 pKa = 3.62GAGEE221 pKa = 4.3SLPPTIDD228 pKa = 2.72QWVYY232 pKa = 10.96NGADD236 pKa = 3.51VNEE239 pKa = 4.94DD240 pKa = 3.58LTFSLSDD247 pKa = 3.44VVDD250 pKa = 4.06AQVTVVDD257 pKa = 3.89VSVPNIFTVSLTDD270 pKa = 3.87LPEE273 pKa = 4.39GTQVQGMVLTSVNGVPTWTATVVVPAGANDD303 pKa = 3.8AAVEE307 pKa = 3.9LALEE311 pKa = 4.37GLLNGIQITPPPNSNEE327 pKa = 3.86NNAEE331 pKa = 3.87DD332 pKa = 4.07AFNFNATLTASAQGSFSVSQTIPDD356 pKa = 3.84TVMVIPVAPVTDD368 pKa = 3.53AAVITVSAVDD378 pKa = 3.39AGEE381 pKa = 4.18NPVDD385 pKa = 3.68GSVSVTIDD393 pKa = 3.22VRR395 pKa = 11.84SNPVDD400 pKa = 3.28GSNGVIVGGLLYY412 pKa = 10.65VQVSATGGGNAGGTLTYY429 pKa = 10.27QGNPAALTPVTNPPGVPAGTYY450 pKa = 9.13YY451 pKa = 10.98LVPVGPSGDD460 pKa = 3.91SVNLVYY466 pKa = 10.29TPPSGTSPGNVIFTALANTQEE487 pKa = 3.97TGAAVVTGSASDD499 pKa = 3.39TATISAVNNGVTVTGFSTPVSAPEE523 pKa = 3.92TDD525 pKa = 3.49GSGLGTAIALPGLMVNLIDD544 pKa = 4.33SDD546 pKa = 3.76GSEE549 pKa = 4.41SIKK552 pKa = 10.83SVTLAGVPVGFLVYY566 pKa = 10.56VGGQLATNAGGNGTSNTWVISNSGTFGEE594 pKa = 4.66VKK596 pKa = 10.05IAPPAHH602 pKa = 6.73WSGEE606 pKa = 4.12LNGLKK611 pKa = 10.36LVVEE615 pKa = 4.49SGEE618 pKa = 4.04NLLAKK623 pKa = 10.35SLEE626 pKa = 4.16EE627 pKa = 3.8FLLPSVTVTPVASGLTLDD645 pKa = 3.51TTQAVGRR652 pKa = 11.84EE653 pKa = 4.05GGIIGLNLNASMTDD667 pKa = 3.38PVAATSTLPDD677 pKa = 3.43SSTEE681 pKa = 4.06TVTVQFKK688 pKa = 11.25GLGEE692 pKa = 3.93YY693 pKa = 10.49AAFYY697 pKa = 10.95VDD699 pKa = 4.04STLLTDD705 pKa = 4.16SGTSGHH711 pKa = 6.45TIGYY715 pKa = 9.78NDD717 pKa = 3.98LTDD720 pKa = 3.65TYY722 pKa = 9.74TITGLTQVEE731 pKa = 4.06MDD733 pKa = 3.3QLGFKK738 pKa = 9.69QAASALVDD746 pKa = 3.37QDD748 pKa = 3.7GAAGTQIEE756 pKa = 4.55VTAWTVEE763 pKa = 4.26SGGGPISAVEE773 pKa = 4.03TADD776 pKa = 2.83ITVNMTVVQPTSGADD791 pKa = 3.24SFIWGGEE798 pKa = 3.67GKK800 pKa = 10.55AIINGAAGDD809 pKa = 3.85DD810 pKa = 3.94TVALRR815 pKa = 11.84LGEE818 pKa = 4.38DD819 pKa = 3.32VSGANLAMGLRR830 pKa = 11.84NIEE833 pKa = 4.08VLDD836 pKa = 4.61LSAQGEE842 pKa = 4.31NAVTNLTATDD852 pKa = 3.76VLSLTGSNHH861 pKa = 6.24ILTINGTGDD870 pKa = 3.87DD871 pKa = 3.99SVQLGGGASAWTAGASSGGYY891 pKa = 8.01TDD893 pKa = 3.69YY894 pKa = 11.35TSGAGASLVTVRR906 pKa = 11.84IDD908 pKa = 3.98DD909 pKa = 5.61DD910 pKa = 3.57ITHH913 pKa = 6.65SYY915 pKa = 8.28VV916 pKa = 2.94
Molecular weight: 92.05 kDa
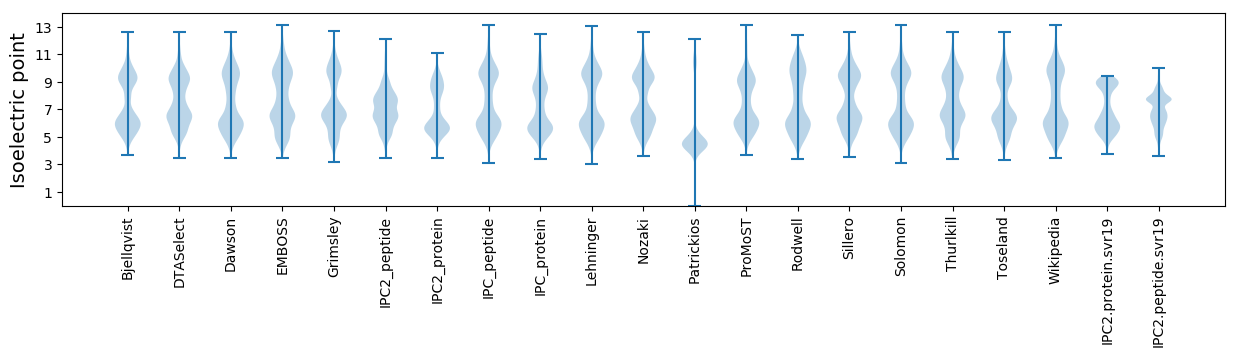
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126ZCD0|A0A126ZCD0_9BURK LuxR family transcriptional regulator OS=Variovorax sp. PAMC 28711 OX=1795631 GN=AX767_06830 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 9.71QASKK9 pKa = 9.04VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.33KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 9.71QASKK9 pKa = 9.04VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1274502 |
32 |
3480 |
321.8 |
34.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.172 ± 0.053 | 0.845 ± 0.011 |
5.384 ± 0.032 | 4.989 ± 0.036 |
3.612 ± 0.026 | 8.516 ± 0.05 |
2.144 ± 0.022 | 4.446 ± 0.026 |
3.384 ± 0.042 | 10.47 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.507 ± 0.018 | 2.626 ± 0.026 |
5.207 ± 0.029 | 3.669 ± 0.026 |
6.698 ± 0.044 | 5.325 ± 0.027 |
5.491 ± 0.035 | 7.928 ± 0.031 |
1.451 ± 0.018 | 2.135 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
