
Candidatus Accumulibacter aalborgensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Accumulibacter
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
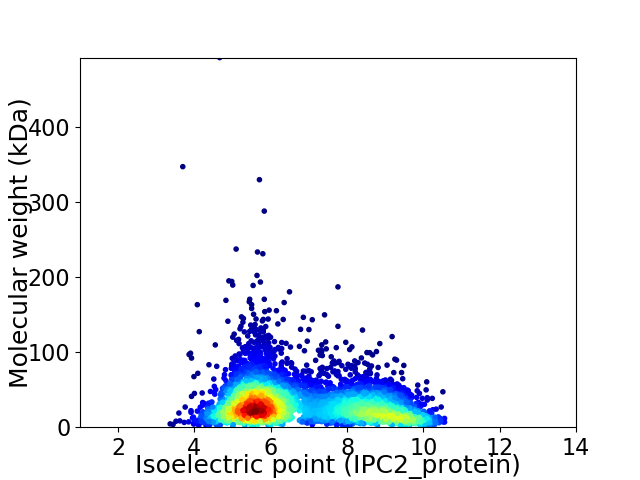
Virtual 2D-PAGE plot for 4556 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A8XIM7|A0A1A8XIM7_9PROT Leucine/isoleucine/valine transporter subunit membrane component of ABC superfamily OS=Candidatus Accumulibacter aalborgensis OX=1860102 GN=livH PE=4 SV=1
MM1 pKa = 7.07ATSTKK6 pKa = 10.19FFDD9 pKa = 4.72DD10 pKa = 4.2PQSFAAVAQVLAGVIEE26 pKa = 4.22PAVPVYY32 pKa = 10.43CGSVPLGNYY41 pKa = 9.04HH42 pKa = 6.34AVEE45 pKa = 4.29VFDD48 pKa = 4.01RR49 pKa = 11.84TPNGSGYY56 pKa = 9.7PGSFVDD62 pKa = 6.23LVANTYY68 pKa = 10.82LRR70 pKa = 11.84DD71 pKa = 3.73TYY73 pKa = 11.12QKK75 pKa = 10.53PDD77 pKa = 3.37GTAGSYY83 pKa = 7.58GTSFAATFSYY93 pKa = 10.54RR94 pKa = 11.84PAAGDD99 pKa = 3.39EE100 pKa = 3.96NLAYY104 pKa = 10.68VPTVDD109 pKa = 5.22RR110 pKa = 11.84GDD112 pKa = 3.27ITIGGGARR120 pKa = 11.84YY121 pKa = 7.87TDD123 pKa = 4.19VITGHH128 pKa = 6.67FGSVANVTSTRR139 pKa = 11.84AFPDD143 pKa = 3.19PTFAFTTIGVSNVFAVQQDD162 pKa = 3.77VEE164 pKa = 4.34LATGVVRR171 pKa = 11.84TAGDD175 pKa = 3.4VLRR178 pKa = 11.84AGTLSSMFSSQQSFDD193 pKa = 3.04ASLILWEE200 pKa = 4.51DD201 pKa = 3.28RR202 pKa = 11.84AGKK205 pKa = 9.18VHH207 pKa = 7.04SLPLSDD213 pKa = 3.38ATPRR217 pKa = 11.84DD218 pKa = 3.49DD219 pKa = 5.49HH220 pKa = 8.36LFDD223 pKa = 3.55QAQEE227 pKa = 3.97LGSWVEE233 pKa = 3.95LVKK236 pKa = 10.88GAGSAWHH243 pKa = 6.99LDD245 pKa = 3.37SPTIRR250 pKa = 11.84LVIDD254 pKa = 3.85NPHH257 pKa = 6.34GLQLGLQGFLASSRR271 pKa = 11.84NPNDD275 pKa = 3.79DD276 pKa = 4.49SLNLWTEE283 pKa = 3.88VLNPSSVLAKK293 pKa = 10.33GLTYY297 pKa = 10.48RR298 pKa = 11.84VDD300 pKa = 4.13FKK302 pKa = 11.1IIAVAPLTDD311 pKa = 3.12NTTYY315 pKa = 9.16VTPFSAKK322 pKa = 10.1LAGTPFSNVTLSGTADD338 pKa = 3.25ANAIGTGGDD347 pKa = 3.37NVIEE351 pKa = 4.55GNSGANTLSGDD362 pKa = 3.76AGNDD366 pKa = 3.42TLTGSAGNDD375 pKa = 3.41TLTGGAGNDD384 pKa = 3.25SFVFADD390 pKa = 4.22HH391 pKa = 7.04GNGLDD396 pKa = 3.87TITDD400 pKa = 4.29FTPGDD405 pKa = 4.0TIDD408 pKa = 3.6VTGVTLVGHH417 pKa = 5.54VTAGDD422 pKa = 3.87GSTVGQGEE430 pKa = 4.35VQFASGTNTLYY441 pKa = 10.4IGTDD445 pKa = 3.31TTSGAEE451 pKa = 4.05VQIKK455 pKa = 10.6LIGSFTAAAFYY466 pKa = 11.19VSGSDD471 pKa = 4.36LSFGPAPTVLSFSPSDD487 pKa = 3.43GATGVATSSNIVLSFSEE504 pKa = 5.0PIAGGTGSILLQTAAGATVEE524 pKa = 4.54TYY526 pKa = 10.81DD527 pKa = 3.5AATSNRR533 pKa = 11.84LTVSGSTLTIDD544 pKa = 3.92PTSSLADD551 pKa = 3.37GTHH554 pKa = 6.3YY555 pKa = 11.0YY556 pKa = 8.6VTLAAGTIKK565 pKa = 10.8DD566 pKa = 3.93LVGNSYY572 pKa = 11.21AGTSTYY578 pKa = 11.42DD579 pKa = 3.38FTTGAAPAPSYY590 pKa = 8.53TVPGTLGNDD599 pKa = 3.55FFIPSAGNNYY609 pKa = 10.3LGGGGNDD616 pKa = 3.68TYY618 pKa = 10.81IISPNTLSGAVTAQITDD635 pKa = 3.81TEE637 pKa = 4.38GSNVIQLVDD646 pKa = 3.55GMTISASSFYY656 pKa = 11.29GNAAQLTLASGAIVQILGASALGYY680 pKa = 9.94QVGANAPAGDD690 pKa = 3.67TAATQTYY697 pKa = 9.38SQFASTLGGSVPTGSTPVSGTAGYY721 pKa = 9.48VVPSGFTQAPALTPAVAGPSYY742 pKa = 9.65TVPGTLGNDD751 pKa = 3.55FFIPSAGNNYY761 pKa = 10.3LGGGGNDD768 pKa = 3.68TYY770 pKa = 10.81IISPNTLSGAVTAQITDD787 pKa = 3.72TEE789 pKa = 4.55GVNVIQWVDD798 pKa = 3.29GMTLTASSFYY808 pKa = 11.15NDD810 pKa = 3.52AAQLTLSTGAKK821 pKa = 9.16VQILGASKK829 pKa = 10.79FNFQLGANAPAGDD842 pKa = 3.77TATSQTYY849 pKa = 10.11AQFASTLGVTLPAAGAAPVSGTPNFVVASSGFDD882 pKa = 3.26VAKK885 pKa = 10.7DD886 pKa = 3.12KK887 pKa = 11.44LVFVNTTDD895 pKa = 3.25QTVYY899 pKa = 10.69TEE901 pKa = 4.78AKK903 pKa = 9.7FKK905 pKa = 10.84ALPGVSIADD914 pKa = 3.37NPFGNNTLVYY924 pKa = 10.33LDD926 pKa = 4.42AASDD930 pKa = 3.76LSDD933 pKa = 3.21GVTLTGIADD942 pKa = 3.81APLSQIVVEE951 pKa = 4.44MMM953 pKa = 3.89
MM1 pKa = 7.07ATSTKK6 pKa = 10.19FFDD9 pKa = 4.72DD10 pKa = 4.2PQSFAAVAQVLAGVIEE26 pKa = 4.22PAVPVYY32 pKa = 10.43CGSVPLGNYY41 pKa = 9.04HH42 pKa = 6.34AVEE45 pKa = 4.29VFDD48 pKa = 4.01RR49 pKa = 11.84TPNGSGYY56 pKa = 9.7PGSFVDD62 pKa = 6.23LVANTYY68 pKa = 10.82LRR70 pKa = 11.84DD71 pKa = 3.73TYY73 pKa = 11.12QKK75 pKa = 10.53PDD77 pKa = 3.37GTAGSYY83 pKa = 7.58GTSFAATFSYY93 pKa = 10.54RR94 pKa = 11.84PAAGDD99 pKa = 3.39EE100 pKa = 3.96NLAYY104 pKa = 10.68VPTVDD109 pKa = 5.22RR110 pKa = 11.84GDD112 pKa = 3.27ITIGGGARR120 pKa = 11.84YY121 pKa = 7.87TDD123 pKa = 4.19VITGHH128 pKa = 6.67FGSVANVTSTRR139 pKa = 11.84AFPDD143 pKa = 3.19PTFAFTTIGVSNVFAVQQDD162 pKa = 3.77VEE164 pKa = 4.34LATGVVRR171 pKa = 11.84TAGDD175 pKa = 3.4VLRR178 pKa = 11.84AGTLSSMFSSQQSFDD193 pKa = 3.04ASLILWEE200 pKa = 4.51DD201 pKa = 3.28RR202 pKa = 11.84AGKK205 pKa = 9.18VHH207 pKa = 7.04SLPLSDD213 pKa = 3.38ATPRR217 pKa = 11.84DD218 pKa = 3.49DD219 pKa = 5.49HH220 pKa = 8.36LFDD223 pKa = 3.55QAQEE227 pKa = 3.97LGSWVEE233 pKa = 3.95LVKK236 pKa = 10.88GAGSAWHH243 pKa = 6.99LDD245 pKa = 3.37SPTIRR250 pKa = 11.84LVIDD254 pKa = 3.85NPHH257 pKa = 6.34GLQLGLQGFLASSRR271 pKa = 11.84NPNDD275 pKa = 3.79DD276 pKa = 4.49SLNLWTEE283 pKa = 3.88VLNPSSVLAKK293 pKa = 10.33GLTYY297 pKa = 10.48RR298 pKa = 11.84VDD300 pKa = 4.13FKK302 pKa = 11.1IIAVAPLTDD311 pKa = 3.12NTTYY315 pKa = 9.16VTPFSAKK322 pKa = 10.1LAGTPFSNVTLSGTADD338 pKa = 3.25ANAIGTGGDD347 pKa = 3.37NVIEE351 pKa = 4.55GNSGANTLSGDD362 pKa = 3.76AGNDD366 pKa = 3.42TLTGSAGNDD375 pKa = 3.41TLTGGAGNDD384 pKa = 3.25SFVFADD390 pKa = 4.22HH391 pKa = 7.04GNGLDD396 pKa = 3.87TITDD400 pKa = 4.29FTPGDD405 pKa = 4.0TIDD408 pKa = 3.6VTGVTLVGHH417 pKa = 5.54VTAGDD422 pKa = 3.87GSTVGQGEE430 pKa = 4.35VQFASGTNTLYY441 pKa = 10.4IGTDD445 pKa = 3.31TTSGAEE451 pKa = 4.05VQIKK455 pKa = 10.6LIGSFTAAAFYY466 pKa = 11.19VSGSDD471 pKa = 4.36LSFGPAPTVLSFSPSDD487 pKa = 3.43GATGVATSSNIVLSFSEE504 pKa = 5.0PIAGGTGSILLQTAAGATVEE524 pKa = 4.54TYY526 pKa = 10.81DD527 pKa = 3.5AATSNRR533 pKa = 11.84LTVSGSTLTIDD544 pKa = 3.92PTSSLADD551 pKa = 3.37GTHH554 pKa = 6.3YY555 pKa = 11.0YY556 pKa = 8.6VTLAAGTIKK565 pKa = 10.8DD566 pKa = 3.93LVGNSYY572 pKa = 11.21AGTSTYY578 pKa = 11.42DD579 pKa = 3.38FTTGAAPAPSYY590 pKa = 8.53TVPGTLGNDD599 pKa = 3.55FFIPSAGNNYY609 pKa = 10.3LGGGGNDD616 pKa = 3.68TYY618 pKa = 10.81IISPNTLSGAVTAQITDD635 pKa = 3.81TEE637 pKa = 4.38GSNVIQLVDD646 pKa = 3.55GMTISASSFYY656 pKa = 11.29GNAAQLTLASGAIVQILGASALGYY680 pKa = 9.94QVGANAPAGDD690 pKa = 3.67TAATQTYY697 pKa = 9.38SQFASTLGGSVPTGSTPVSGTAGYY721 pKa = 9.48VVPSGFTQAPALTPAVAGPSYY742 pKa = 9.65TVPGTLGNDD751 pKa = 3.55FFIPSAGNNYY761 pKa = 10.3LGGGGNDD768 pKa = 3.68TYY770 pKa = 10.81IISPNTLSGAVTAQITDD787 pKa = 3.72TEE789 pKa = 4.55GVNVIQWVDD798 pKa = 3.29GMTLTASSFYY808 pKa = 11.15NDD810 pKa = 3.52AAQLTLSTGAKK821 pKa = 9.16VQILGASKK829 pKa = 10.79FNFQLGANAPAGDD842 pKa = 3.77TATSQTYY849 pKa = 10.11AQFASTLGVTLPAAGAAPVSGTPNFVVASSGFDD882 pKa = 3.26VAKK885 pKa = 10.7DD886 pKa = 3.12KK887 pKa = 11.44LVFVNTTDD895 pKa = 3.25QTVYY899 pKa = 10.69TEE901 pKa = 4.78AKK903 pKa = 9.7FKK905 pKa = 10.84ALPGVSIADD914 pKa = 3.37NPFGNNTLVYY924 pKa = 10.33LDD926 pKa = 4.42AASDD930 pKa = 3.76LSDD933 pKa = 3.21GVTLTGIADD942 pKa = 3.81APLSQIVVEE951 pKa = 4.44MMM953 pKa = 3.89
Molecular weight: 96.76 kDa
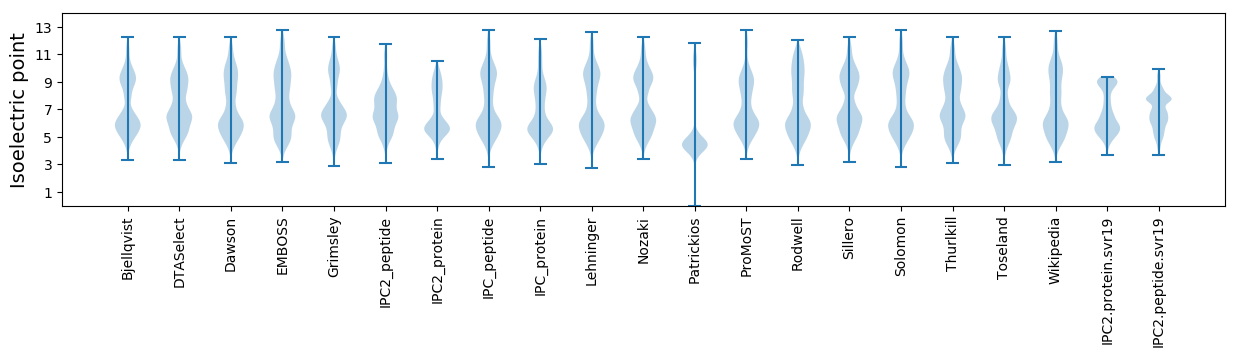
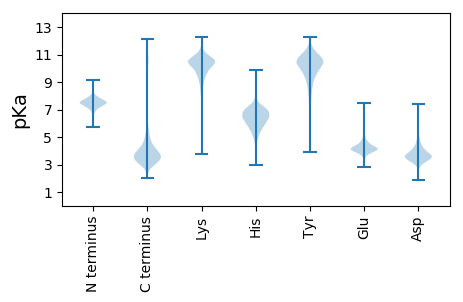
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A8XJ88|A0A1A8XJ88_9PROT Uncharacterized protein OS=Candidatus Accumulibacter aalborgensis OX=1860102 GN=ACCAA_180043 PE=4 SV=1
MM1 pKa = 8.09DD2 pKa = 3.92GRR4 pKa = 11.84RR5 pKa = 11.84TRR7 pKa = 11.84SLEE10 pKa = 3.88RR11 pKa = 11.84TILAATSASFSSTPLRR27 pKa = 11.84EE28 pKa = 4.1CRR30 pKa = 11.84LDD32 pKa = 3.43ALGDD36 pKa = 3.77VEE38 pKa = 4.08ICKK41 pKa = 10.23PNRR44 pKa = 11.84KK45 pKa = 8.94EE46 pKa = 3.83NCCPNKK52 pKa = 9.87LHH54 pKa = 6.82VYY56 pKa = 10.01IIRR59 pKa = 11.84HH60 pKa = 5.41GLLIEE65 pKa = 4.83DD66 pKa = 4.15PSVVRR71 pKa = 11.84MHH73 pKa = 7.19RR74 pKa = 11.84AHH76 pKa = 7.46ASRR79 pKa = 11.84GQLHH83 pKa = 6.66RR84 pKa = 11.84GVSSNIRR91 pKa = 11.84TVKK94 pKa = 10.37QLLGQLARR102 pKa = 11.84NILIEE107 pKa = 4.08TLRR110 pKa = 11.84TSQRR114 pKa = 11.84LARR117 pKa = 11.84FAVCFSGLVRR127 pKa = 11.84ACAGSCKK134 pKa = 10.52NKK136 pKa = 9.73FQRR139 pKa = 11.84LQQSAVAWATAVAVAEE155 pKa = 4.8HH156 pKa = 6.03YY157 pKa = 8.98WGLVNNEE164 pKa = 3.2WRR166 pKa = 11.84VPCSRR171 pKa = 11.84VNPCGLARR179 pKa = 4.32
MM1 pKa = 8.09DD2 pKa = 3.92GRR4 pKa = 11.84RR5 pKa = 11.84TRR7 pKa = 11.84SLEE10 pKa = 3.88RR11 pKa = 11.84TILAATSASFSSTPLRR27 pKa = 11.84EE28 pKa = 4.1CRR30 pKa = 11.84LDD32 pKa = 3.43ALGDD36 pKa = 3.77VEE38 pKa = 4.08ICKK41 pKa = 10.23PNRR44 pKa = 11.84KK45 pKa = 8.94EE46 pKa = 3.83NCCPNKK52 pKa = 9.87LHH54 pKa = 6.82VYY56 pKa = 10.01IIRR59 pKa = 11.84HH60 pKa = 5.41GLLIEE65 pKa = 4.83DD66 pKa = 4.15PSVVRR71 pKa = 11.84MHH73 pKa = 7.19RR74 pKa = 11.84AHH76 pKa = 7.46ASRR79 pKa = 11.84GQLHH83 pKa = 6.66RR84 pKa = 11.84GVSSNIRR91 pKa = 11.84TVKK94 pKa = 10.37QLLGQLARR102 pKa = 11.84NILIEE107 pKa = 4.08TLRR110 pKa = 11.84TSQRR114 pKa = 11.84LARR117 pKa = 11.84FAVCFSGLVRR127 pKa = 11.84ACAGSCKK134 pKa = 10.52NKK136 pKa = 9.73FQRR139 pKa = 11.84LQQSAVAWATAVAVAEE155 pKa = 4.8HH156 pKa = 6.03YY157 pKa = 8.98WGLVNNEE164 pKa = 3.2WRR166 pKa = 11.84VPCSRR171 pKa = 11.84VNPCGLARR179 pKa = 4.32
Molecular weight: 20.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1422198 |
19 |
4715 |
312.2 |
34.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.09 ± 0.052 | 1.075 ± 0.014 |
5.487 ± 0.027 | 5.54 ± 0.038 |
3.683 ± 0.026 | 7.964 ± 0.037 |
2.248 ± 0.022 | 4.765 ± 0.03 |
3.173 ± 0.035 | 11.119 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.189 ± 0.017 | 2.664 ± 0.029 |
5.054 ± 0.025 | 3.769 ± 0.024 |
7.39 ± 0.042 | 5.696 ± 0.035 |
5.027 ± 0.041 | 7.341 ± 0.031 |
1.402 ± 0.015 | 2.324 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
