
Barley virus G
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
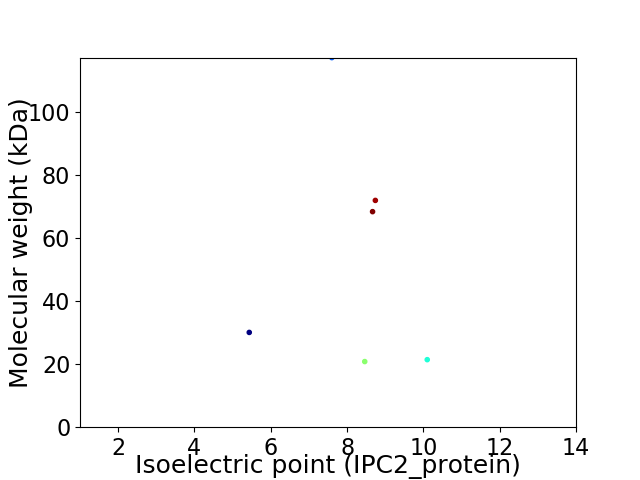
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A165ELR9|A0A165ELR9_9LUTE Serine protease OS=Barley virus G OX=1825924 PE=4 SV=1
MM1 pKa = 7.61RR2 pKa = 11.84CAINFDD8 pKa = 3.7GTLEE12 pKa = 4.05FSDD15 pKa = 3.72IPRR18 pKa = 11.84DD19 pKa = 3.44LSVRR23 pKa = 11.84LAEE26 pKa = 4.45FVSATITIAFCALQPEE42 pKa = 4.63YY43 pKa = 10.68DD44 pKa = 3.54AEE46 pKa = 4.16ALFRR50 pKa = 11.84SVLTVLPLMLRR61 pKa = 11.84PSPYY65 pKa = 8.11TRR67 pKa = 11.84GIARR71 pKa = 11.84EE72 pKa = 4.02AWSSFARR79 pKa = 11.84HH80 pKa = 6.1ALRR83 pKa = 11.84TGGRR87 pKa = 11.84ISLPPEE93 pKa = 4.0FNDD96 pKa = 4.88NSDD99 pKa = 3.26EE100 pKa = 4.08PAALFRR106 pKa = 11.84RR107 pKa = 11.84FLQRR111 pKa = 11.84VDD113 pKa = 2.98ARR115 pKa = 11.84AYY117 pKa = 9.53AHH119 pKa = 6.55ALKK122 pKa = 10.57GRR124 pKa = 11.84EE125 pKa = 4.07EE126 pKa = 4.18NFFTSLRR133 pKa = 11.84SFQLKK138 pKa = 10.53LEE140 pKa = 4.41SFCRR144 pKa = 11.84RR145 pKa = 11.84AEE147 pKa = 4.21DD148 pKa = 3.7PDD150 pKa = 4.57RR151 pKa = 11.84YY152 pKa = 8.63PQRR155 pKa = 11.84RR156 pKa = 11.84HH157 pKa = 6.33IEE159 pKa = 4.02VLSGIVRR166 pKa = 11.84EE167 pKa = 4.04AHH169 pKa = 6.19NNGEE173 pKa = 4.04SIYY176 pKa = 10.83RR177 pKa = 11.84HH178 pKa = 6.15FPDD181 pKa = 4.63ADD183 pKa = 4.04NIHH186 pKa = 6.96LEE188 pKa = 3.95LLDD191 pKa = 4.27LGRR194 pKa = 11.84SLEE197 pKa = 4.04FDD199 pKa = 4.03FPHH202 pKa = 6.28QAVHH206 pKa = 6.59PSDD209 pKa = 4.85LISRR213 pKa = 11.84LFVYY217 pKa = 7.84MHH219 pKa = 6.81NALGPFGLQDD229 pKa = 3.5FWRR232 pKa = 11.84VADD235 pKa = 4.17LSDD238 pKa = 3.62FFVIHH243 pKa = 7.09SDD245 pKa = 3.35VQYY248 pKa = 11.0LQDD251 pKa = 3.9SYY253 pKa = 11.27IQKK256 pKa = 10.17EE257 pKa = 4.04LADD260 pKa = 3.66VV261 pKa = 3.7
MM1 pKa = 7.61RR2 pKa = 11.84CAINFDD8 pKa = 3.7GTLEE12 pKa = 4.05FSDD15 pKa = 3.72IPRR18 pKa = 11.84DD19 pKa = 3.44LSVRR23 pKa = 11.84LAEE26 pKa = 4.45FVSATITIAFCALQPEE42 pKa = 4.63YY43 pKa = 10.68DD44 pKa = 3.54AEE46 pKa = 4.16ALFRR50 pKa = 11.84SVLTVLPLMLRR61 pKa = 11.84PSPYY65 pKa = 8.11TRR67 pKa = 11.84GIARR71 pKa = 11.84EE72 pKa = 4.02AWSSFARR79 pKa = 11.84HH80 pKa = 6.1ALRR83 pKa = 11.84TGGRR87 pKa = 11.84ISLPPEE93 pKa = 4.0FNDD96 pKa = 4.88NSDD99 pKa = 3.26EE100 pKa = 4.08PAALFRR106 pKa = 11.84RR107 pKa = 11.84FLQRR111 pKa = 11.84VDD113 pKa = 2.98ARR115 pKa = 11.84AYY117 pKa = 9.53AHH119 pKa = 6.55ALKK122 pKa = 10.57GRR124 pKa = 11.84EE125 pKa = 4.07EE126 pKa = 4.18NFFTSLRR133 pKa = 11.84SFQLKK138 pKa = 10.53LEE140 pKa = 4.41SFCRR144 pKa = 11.84RR145 pKa = 11.84AEE147 pKa = 4.21DD148 pKa = 3.7PDD150 pKa = 4.57RR151 pKa = 11.84YY152 pKa = 8.63PQRR155 pKa = 11.84RR156 pKa = 11.84HH157 pKa = 6.33IEE159 pKa = 4.02VLSGIVRR166 pKa = 11.84EE167 pKa = 4.04AHH169 pKa = 6.19NNGEE173 pKa = 4.04SIYY176 pKa = 10.83RR177 pKa = 11.84HH178 pKa = 6.15FPDD181 pKa = 4.63ADD183 pKa = 4.04NIHH186 pKa = 6.96LEE188 pKa = 3.95LLDD191 pKa = 4.27LGRR194 pKa = 11.84SLEE197 pKa = 4.04FDD199 pKa = 4.03FPHH202 pKa = 6.28QAVHH206 pKa = 6.59PSDD209 pKa = 4.85LISRR213 pKa = 11.84LFVYY217 pKa = 7.84MHH219 pKa = 6.81NALGPFGLQDD229 pKa = 3.5FWRR232 pKa = 11.84VADD235 pKa = 4.17LSDD238 pKa = 3.62FFVIHH243 pKa = 7.09SDD245 pKa = 3.35VQYY248 pKa = 11.0LQDD251 pKa = 3.9SYY253 pKa = 11.27IQKK256 pKa = 10.17EE257 pKa = 4.04LADD260 pKa = 3.66VV261 pKa = 3.7
Molecular weight: 30.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A165ELU0|A0A165ELU0_9LUTE Movement protein OS=Barley virus G OX=1825924 PE=3 SV=1
MM1 pKa = 6.87NTGGRR6 pKa = 11.84NGRR9 pKa = 11.84RR10 pKa = 11.84TRR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VRR18 pKa = 11.84PASRR22 pKa = 11.84TQPVVVVAAGQRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84RR44 pKa = 11.84TGNTSGGSGIRR55 pKa = 11.84RR56 pKa = 11.84GSRR59 pKa = 11.84EE60 pKa = 3.74TFVFSKK66 pKa = 11.02DD67 pKa = 3.43SLTGNASGKK76 pKa = 8.55LTFGASLSEE85 pKa = 4.14CAAFSSGILKK95 pKa = 10.26AYY97 pKa = 10.28HH98 pKa = 6.69EE99 pKa = 4.41YY100 pKa = 10.53KK101 pKa = 10.1ISKK104 pKa = 7.96VTLEE108 pKa = 4.8FISEE112 pKa = 4.08ASSQSEE118 pKa = 3.81GSIAYY123 pKa = 9.56EE124 pKa = 3.64LDD126 pKa = 3.21PHH128 pKa = 6.71NKK130 pKa = 9.64LSALSSTINKK140 pKa = 9.53FSIVKK145 pKa = 9.56GGKK148 pKa = 8.18RR149 pKa = 11.84TFTSNQIGGGVWRR162 pKa = 11.84DD163 pKa = 3.34STEE166 pKa = 3.77DD167 pKa = 3.26QFAILYY173 pKa = 9.16KK174 pKa = 11.02GNGKK178 pKa = 10.17SSIAGSFRR186 pKa = 11.84VTMDD190 pKa = 3.75VLTQNPKK197 pKa = 10.33
MM1 pKa = 6.87NTGGRR6 pKa = 11.84NGRR9 pKa = 11.84RR10 pKa = 11.84TRR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VRR18 pKa = 11.84PASRR22 pKa = 11.84TQPVVVVAAGQRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84RR44 pKa = 11.84TGNTSGGSGIRR55 pKa = 11.84RR56 pKa = 11.84GSRR59 pKa = 11.84EE60 pKa = 3.74TFVFSKK66 pKa = 11.02DD67 pKa = 3.43SLTGNASGKK76 pKa = 8.55LTFGASLSEE85 pKa = 4.14CAAFSSGILKK95 pKa = 10.26AYY97 pKa = 10.28HH98 pKa = 6.69EE99 pKa = 4.41YY100 pKa = 10.53KK101 pKa = 10.1ISKK104 pKa = 7.96VTLEE108 pKa = 4.8FISEE112 pKa = 4.08ASSQSEE118 pKa = 3.81GSIAYY123 pKa = 9.56EE124 pKa = 3.64LDD126 pKa = 3.21PHH128 pKa = 6.71NKK130 pKa = 9.64LSALSSTINKK140 pKa = 9.53FSIVKK145 pKa = 9.56GGKK148 pKa = 8.18RR149 pKa = 11.84TFTSNQIGGGVWRR162 pKa = 11.84DD163 pKa = 3.34STEE166 pKa = 3.77DD167 pKa = 3.26QFAILYY173 pKa = 9.16KK174 pKa = 11.02GNGKK178 pKa = 10.17SSIAGSFRR186 pKa = 11.84VTMDD190 pKa = 3.75VLTQNPKK197 pKa = 10.33
Molecular weight: 21.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2961 |
185 |
1043 |
493.5 |
55.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.856 ± 0.329 | 1.621 ± 0.398 |
4.593 ± 0.492 | 5.505 ± 0.373 |
4.154 ± 0.484 | 6.788 ± 0.764 |
1.486 ± 0.292 | 4.357 ± 0.359 |
4.998 ± 0.568 | 9.355 ± 1.368 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.689 ± 0.172 | 4.222 ± 0.239 |
5.674 ± 0.684 | 4.694 ± 0.357 |
7.295 ± 0.829 | 10.199 ± 0.972 |
6.011 ± 0.529 | 5.775 ± 0.159 |
1.52 ± 0.227 | 3.175 ± 0.252 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
