
Idiomarina loihiensis (strain ATCC BAA-735 / DSM 15497 / L2-TR)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Idiomarinaceae; Idiomarina; Idiomarina loihiensis
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
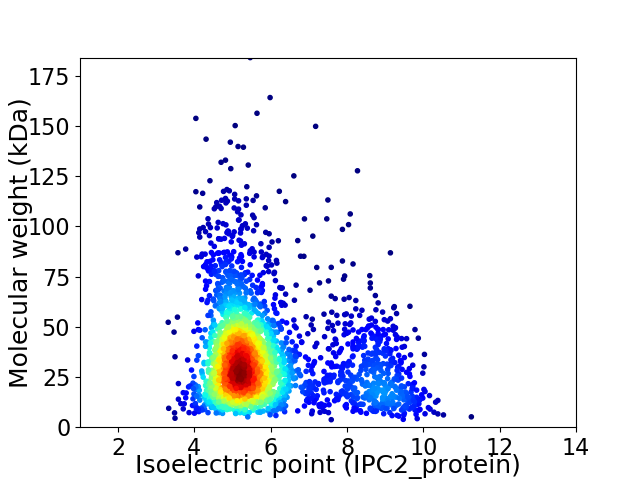
Virtual 2D-PAGE plot for 2608 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5QZG3|Q5QZG3_IDILO Possible Zn-finger protein OS=Idiomarina loihiensis (strain ATCC BAA-735 / DSM 15497 / L2-TR) OX=283942 GN=IL2600 PE=4 SV=1
MM1 pKa = 7.86FYY3 pKa = 10.39RR4 pKa = 11.84NCFAASLLVLACSQLTACNSDD25 pKa = 3.31SSSEE29 pKa = 4.21SSDD32 pKa = 3.33HH33 pKa = 6.01QEE35 pKa = 3.98SGIWQPKK42 pKa = 9.32PGTSWHH48 pKa = 5.94WQLEE52 pKa = 4.4NYY54 pKa = 9.93DD55 pKa = 4.16NLDD58 pKa = 3.16ISKK61 pKa = 10.4DD62 pKa = 3.46AEE64 pKa = 4.13AFDD67 pKa = 3.83IDD69 pKa = 4.5LFEE72 pKa = 4.47GAEE75 pKa = 4.31GGDD78 pKa = 3.73DD79 pKa = 4.91SIISSLKK86 pKa = 10.54DD87 pKa = 2.88NGKK90 pKa = 9.74RR91 pKa = 11.84VICYY95 pKa = 8.37FSAGTRR101 pKa = 11.84EE102 pKa = 4.19DD103 pKa = 3.26WRR105 pKa = 11.84PDD107 pKa = 2.86ATEE110 pKa = 3.91FSEE113 pKa = 5.34DD114 pKa = 3.17AVIANGEE121 pKa = 4.03MADD124 pKa = 3.5WPGEE128 pKa = 3.9VWLDD132 pKa = 3.45INNEE136 pKa = 3.71AVLNEE141 pKa = 4.11NIKK144 pKa = 10.57PIMEE148 pKa = 4.43ARR150 pKa = 11.84LDD152 pKa = 3.8LAQSAGCDD160 pKa = 3.03AVEE163 pKa = 4.38PDD165 pKa = 4.54NVDD168 pKa = 3.28GYY170 pKa = 10.95INTDD174 pKa = 3.19EE175 pKa = 4.3TKK177 pKa = 11.06GIITYY182 pKa = 10.6DD183 pKa = 3.41DD184 pKa = 3.34QLNYY188 pKa = 10.95NKK190 pKa = 9.73WLANAAHH197 pKa = 6.33SRR199 pKa = 11.84GLSIGLKK206 pKa = 10.24NDD208 pKa = 2.84VDD210 pKa = 3.93QLNEE214 pKa = 4.07LVNDD218 pKa = 3.79FDD220 pKa = 4.47FAVNEE225 pKa = 3.67QCYY228 pKa = 10.04AYY230 pKa = 10.5GNEE233 pKa = 4.12CVSYY237 pKa = 11.22EE238 pKa = 4.39DD239 pKa = 4.71TFLKK243 pKa = 10.51NNKK246 pKa = 9.19AVFVQEE252 pKa = 4.13YY253 pKa = 9.9YY254 pKa = 10.8EE255 pKa = 4.96DD256 pKa = 4.16GSEE259 pKa = 4.6GEE261 pKa = 3.95ISQQEE266 pKa = 4.38FEE268 pKa = 5.44SNACSYY274 pKa = 10.61FLDD277 pKa = 3.84VGISALWKK285 pKa = 10.45EE286 pKa = 4.36GFNLDD291 pKa = 3.69GEE293 pKa = 4.79NVLSCSEE300 pKa = 3.96
MM1 pKa = 7.86FYY3 pKa = 10.39RR4 pKa = 11.84NCFAASLLVLACSQLTACNSDD25 pKa = 3.31SSSEE29 pKa = 4.21SSDD32 pKa = 3.33HH33 pKa = 6.01QEE35 pKa = 3.98SGIWQPKK42 pKa = 9.32PGTSWHH48 pKa = 5.94WQLEE52 pKa = 4.4NYY54 pKa = 9.93DD55 pKa = 4.16NLDD58 pKa = 3.16ISKK61 pKa = 10.4DD62 pKa = 3.46AEE64 pKa = 4.13AFDD67 pKa = 3.83IDD69 pKa = 4.5LFEE72 pKa = 4.47GAEE75 pKa = 4.31GGDD78 pKa = 3.73DD79 pKa = 4.91SIISSLKK86 pKa = 10.54DD87 pKa = 2.88NGKK90 pKa = 9.74RR91 pKa = 11.84VICYY95 pKa = 8.37FSAGTRR101 pKa = 11.84EE102 pKa = 4.19DD103 pKa = 3.26WRR105 pKa = 11.84PDD107 pKa = 2.86ATEE110 pKa = 3.91FSEE113 pKa = 5.34DD114 pKa = 3.17AVIANGEE121 pKa = 4.03MADD124 pKa = 3.5WPGEE128 pKa = 3.9VWLDD132 pKa = 3.45INNEE136 pKa = 3.71AVLNEE141 pKa = 4.11NIKK144 pKa = 10.57PIMEE148 pKa = 4.43ARR150 pKa = 11.84LDD152 pKa = 3.8LAQSAGCDD160 pKa = 3.03AVEE163 pKa = 4.38PDD165 pKa = 4.54NVDD168 pKa = 3.28GYY170 pKa = 10.95INTDD174 pKa = 3.19EE175 pKa = 4.3TKK177 pKa = 11.06GIITYY182 pKa = 10.6DD183 pKa = 3.41DD184 pKa = 3.34QLNYY188 pKa = 10.95NKK190 pKa = 9.73WLANAAHH197 pKa = 6.33SRR199 pKa = 11.84GLSIGLKK206 pKa = 10.24NDD208 pKa = 2.84VDD210 pKa = 3.93QLNEE214 pKa = 4.07LVNDD218 pKa = 3.79FDD220 pKa = 4.47FAVNEE225 pKa = 3.67QCYY228 pKa = 10.04AYY230 pKa = 10.5GNEE233 pKa = 4.12CVSYY237 pKa = 11.22EE238 pKa = 4.39DD239 pKa = 4.71TFLKK243 pKa = 10.51NNKK246 pKa = 9.19AVFVQEE252 pKa = 4.13YY253 pKa = 9.9YY254 pKa = 10.8EE255 pKa = 4.96DD256 pKa = 4.16GSEE259 pKa = 4.6GEE261 pKa = 3.95ISQQEE266 pKa = 4.38FEE268 pKa = 5.44SNACSYY274 pKa = 10.61FLDD277 pKa = 3.84VGISALWKK285 pKa = 10.45EE286 pKa = 4.36GFNLDD291 pKa = 3.69GEE293 pKa = 4.79NVLSCSEE300 pKa = 3.96
Molecular weight: 33.48 kDa
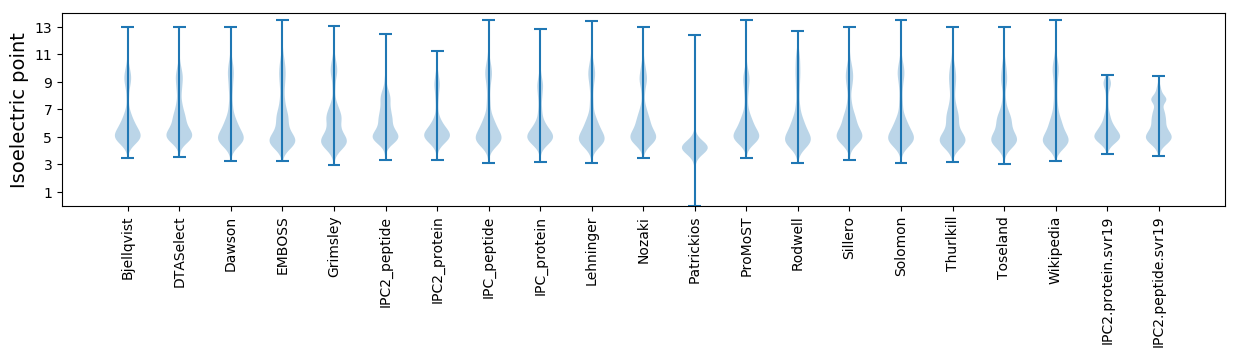
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5QZK6|Q5QZK6_IDILO Probable p-aminobenzoyl-glutamate transporter OS=Idiomarina loihiensis (strain ATCC BAA-735 / DSM 15497 / L2-TR) OX=283942 GN=abgT_2 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
862909 |
34 |
1615 |
330.9 |
36.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.775 ± 0.048 | 0.845 ± 0.015 |
5.78 ± 0.035 | 6.976 ± 0.047 |
3.966 ± 0.034 | 6.911 ± 0.05 |
2.153 ± 0.026 | 5.736 ± 0.037 |
4.989 ± 0.038 | 10.341 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.462 ± 0.023 | 4.024 ± 0.032 |
4.005 ± 0.026 | 4.834 ± 0.048 |
5.193 ± 0.04 | 6.493 ± 0.038 |
5.092 ± 0.03 | 7.08 ± 0.043 |
1.362 ± 0.019 | 2.985 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
