
Maize-associated pteridovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Mayoviridae; Pteridovirus; Maize associated pteridovirus
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
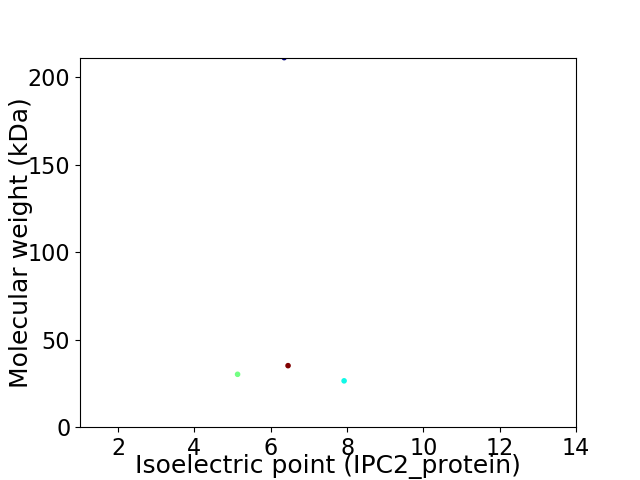
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9D1K2|A0A3Q9D1K2_9VIRU RNA helicase OS=Maize-associated pteridovirus OX=2497338 PE=4 SV=1
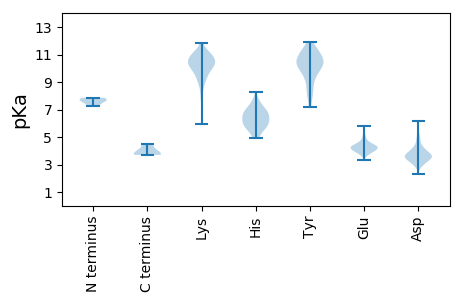
MM1 pKa = 7.24FPEE4 pKa = 5.81SIAQMLKK11 pKa = 9.63SRR13 pKa = 11.84VRR15 pKa = 11.84RR16 pKa = 11.84SASAAVFAAAAASADD31 pKa = 3.53AVAGTLRR38 pKa = 11.84EE39 pKa = 4.13FEE41 pKa = 3.99ISLARR46 pKa = 11.84DD47 pKa = 3.47LDD49 pKa = 3.82AVEE52 pKa = 4.54YY53 pKa = 10.45EE54 pKa = 4.39SFVTDD59 pKa = 3.7SEE61 pKa = 4.77GVPAMVDD68 pKa = 3.2DD69 pKa = 5.61APITRR74 pKa = 11.84CGPARR79 pKa = 11.84YY80 pKa = 8.74VWGVAALSGYY90 pKa = 8.03TPTVIGRR97 pKa = 11.84LFRR100 pKa = 11.84HH101 pKa = 5.91KK102 pKa = 10.56RR103 pKa = 11.84PMGISGSVEE112 pKa = 3.72PLSIDD117 pKa = 4.15DD118 pKa = 3.97YY119 pKa = 11.57TDD121 pKa = 2.88VNYY124 pKa = 10.61CVEE127 pKa = 3.85HH128 pKa = 7.37DD129 pKa = 3.7IFYY132 pKa = 10.43EE133 pKa = 4.3CTQEE137 pKa = 4.32SVCMRR142 pKa = 11.84CVDD145 pKa = 3.84AGVMFTPNVLDD156 pKa = 4.2RR157 pKa = 11.84DD158 pKa = 4.05LVPYY162 pKa = 9.53TRR164 pKa = 11.84IRR166 pKa = 11.84GSEE169 pKa = 4.46LIPTYY174 pKa = 10.69VGRR177 pKa = 11.84EE178 pKa = 3.96YY179 pKa = 11.34LNGIPRR185 pKa = 11.84HH186 pKa = 5.15HH187 pKa = 7.63WYY189 pKa = 10.6LSSQSVLTCACGSSDD204 pKa = 4.98HH205 pKa = 6.55EE206 pKa = 4.6FEE208 pKa = 4.91FDD210 pKa = 3.48GFPPIWFKK218 pKa = 11.15PPASFAYY225 pKa = 10.36ALFPTSLRR233 pKa = 11.84EE234 pKa = 3.91EE235 pKa = 5.03GEE237 pKa = 4.18TSSLVLSSFGYY248 pKa = 9.92IVGRR252 pKa = 11.84YY253 pKa = 8.11GKK255 pKa = 9.27RR256 pKa = 11.84QQSQVPAGQVARR268 pKa = 11.84SSVSLGLL275 pKa = 3.7
MM1 pKa = 7.24FPEE4 pKa = 5.81SIAQMLKK11 pKa = 9.63SRR13 pKa = 11.84VRR15 pKa = 11.84RR16 pKa = 11.84SASAAVFAAAAASADD31 pKa = 3.53AVAGTLRR38 pKa = 11.84EE39 pKa = 4.13FEE41 pKa = 3.99ISLARR46 pKa = 11.84DD47 pKa = 3.47LDD49 pKa = 3.82AVEE52 pKa = 4.54YY53 pKa = 10.45EE54 pKa = 4.39SFVTDD59 pKa = 3.7SEE61 pKa = 4.77GVPAMVDD68 pKa = 3.2DD69 pKa = 5.61APITRR74 pKa = 11.84CGPARR79 pKa = 11.84YY80 pKa = 8.74VWGVAALSGYY90 pKa = 8.03TPTVIGRR97 pKa = 11.84LFRR100 pKa = 11.84HH101 pKa = 5.91KK102 pKa = 10.56RR103 pKa = 11.84PMGISGSVEE112 pKa = 3.72PLSIDD117 pKa = 4.15DD118 pKa = 3.97YY119 pKa = 11.57TDD121 pKa = 2.88VNYY124 pKa = 10.61CVEE127 pKa = 3.85HH128 pKa = 7.37DD129 pKa = 3.7IFYY132 pKa = 10.43EE133 pKa = 4.3CTQEE137 pKa = 4.32SVCMRR142 pKa = 11.84CVDD145 pKa = 3.84AGVMFTPNVLDD156 pKa = 4.2RR157 pKa = 11.84DD158 pKa = 4.05LVPYY162 pKa = 9.53TRR164 pKa = 11.84IRR166 pKa = 11.84GSEE169 pKa = 4.46LIPTYY174 pKa = 10.69VGRR177 pKa = 11.84EE178 pKa = 3.96YY179 pKa = 11.34LNGIPRR185 pKa = 11.84HH186 pKa = 5.15HH187 pKa = 7.63WYY189 pKa = 10.6LSSQSVLTCACGSSDD204 pKa = 4.98HH205 pKa = 6.55EE206 pKa = 4.6FEE208 pKa = 4.91FDD210 pKa = 3.48GFPPIWFKK218 pKa = 11.15PPASFAYY225 pKa = 10.36ALFPTSLRR233 pKa = 11.84EE234 pKa = 3.91EE235 pKa = 5.03GEE237 pKa = 4.18TSSLVLSSFGYY248 pKa = 9.92IVGRR252 pKa = 11.84YY253 pKa = 8.11GKK255 pKa = 9.27RR256 pKa = 11.84QQSQVPAGQVARR268 pKa = 11.84SSVSLGLL275 pKa = 3.7
Molecular weight: 30.22 kDa
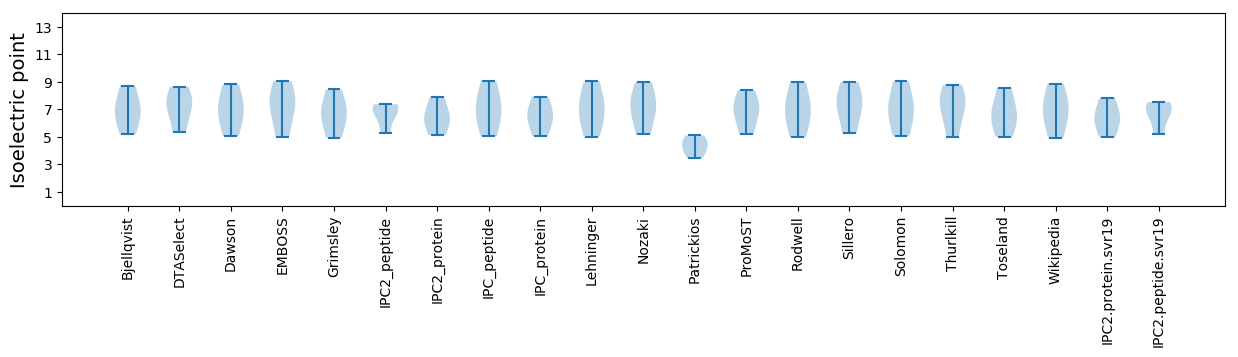
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q9D1K1|A0A3Q9D1K1_9VIRU ORF2 OS=Maize-associated pteridovirus OX=2497338 PE=4 SV=1
MM1 pKa = 7.86ASGSKK6 pKa = 10.34AKK8 pKa = 10.8SPLDD12 pKa = 3.5KK13 pKa = 11.26LLDD16 pKa = 3.47QALALGFDD24 pKa = 4.49VKK26 pKa = 10.93RR27 pKa = 11.84KK28 pKa = 10.12GNDD31 pKa = 3.29GASFSQRR38 pKa = 11.84DD39 pKa = 3.46GASKK43 pKa = 10.96VKK45 pKa = 10.12ILKK48 pKa = 8.86TFLANQRR55 pKa = 11.84KK56 pKa = 9.03KK57 pKa = 10.86NKK59 pKa = 9.43GVAADD64 pKa = 3.72VPGQSHH70 pKa = 5.58VANPARR76 pKa = 11.84VVQEE80 pKa = 3.41QSARR84 pKa = 11.84LEE86 pKa = 4.29WIPLISKK93 pKa = 7.62WTALDD98 pKa = 3.25HH99 pKa = 5.6TKK101 pKa = 10.37EE102 pKa = 4.09YY103 pKa = 10.71QRR105 pKa = 11.84LPMTSEE111 pKa = 3.53IQKK114 pKa = 10.27EE115 pKa = 4.35KK116 pKa = 9.12VHH118 pKa = 6.69KK119 pKa = 10.56LVFSLKK125 pKa = 9.82IAGSVALTEE134 pKa = 4.26PVAKK138 pKa = 9.95VVPVFNTNIKK148 pKa = 10.82GDD150 pKa = 4.05DD151 pKa = 3.68LKK153 pKa = 10.33TASVEE158 pKa = 4.21SDD160 pKa = 3.65FSSLIISNQNGEE172 pKa = 4.21SPYY175 pKa = 10.3VLEE178 pKa = 4.38FAEE181 pKa = 4.69PCTTEE186 pKa = 5.0LIGAKK191 pKa = 10.01LSMVVATGNTEE202 pKa = 4.22VGTEE206 pKa = 3.68WLYY209 pKa = 11.06YY210 pKa = 8.38KK211 pKa = 10.61AWMSAEE217 pKa = 4.43VASHH221 pKa = 7.03ASPNLEE227 pKa = 4.0NAFVAALLDD236 pKa = 3.96SCLSRR241 pKa = 11.84AFNN244 pKa = 3.89
MM1 pKa = 7.86ASGSKK6 pKa = 10.34AKK8 pKa = 10.8SPLDD12 pKa = 3.5KK13 pKa = 11.26LLDD16 pKa = 3.47QALALGFDD24 pKa = 4.49VKK26 pKa = 10.93RR27 pKa = 11.84KK28 pKa = 10.12GNDD31 pKa = 3.29GASFSQRR38 pKa = 11.84DD39 pKa = 3.46GASKK43 pKa = 10.96VKK45 pKa = 10.12ILKK48 pKa = 8.86TFLANQRR55 pKa = 11.84KK56 pKa = 9.03KK57 pKa = 10.86NKK59 pKa = 9.43GVAADD64 pKa = 3.72VPGQSHH70 pKa = 5.58VANPARR76 pKa = 11.84VVQEE80 pKa = 3.41QSARR84 pKa = 11.84LEE86 pKa = 4.29WIPLISKK93 pKa = 7.62WTALDD98 pKa = 3.25HH99 pKa = 5.6TKK101 pKa = 10.37EE102 pKa = 4.09YY103 pKa = 10.71QRR105 pKa = 11.84LPMTSEE111 pKa = 3.53IQKK114 pKa = 10.27EE115 pKa = 4.35KK116 pKa = 9.12VHH118 pKa = 6.69KK119 pKa = 10.56LVFSLKK125 pKa = 9.82IAGSVALTEE134 pKa = 4.26PVAKK138 pKa = 9.95VVPVFNTNIKK148 pKa = 10.82GDD150 pKa = 4.05DD151 pKa = 3.68LKK153 pKa = 10.33TASVEE158 pKa = 4.21SDD160 pKa = 3.65FSSLIISNQNGEE172 pKa = 4.21SPYY175 pKa = 10.3VLEE178 pKa = 4.38FAEE181 pKa = 4.69PCTTEE186 pKa = 5.0LIGAKK191 pKa = 10.01LSMVVATGNTEE202 pKa = 4.22VGTEE206 pKa = 3.68WLYY209 pKa = 11.06YY210 pKa = 8.38KK211 pKa = 10.61AWMSAEE217 pKa = 4.43VASHH221 pKa = 7.03ASPNLEE227 pKa = 4.0NAFVAALLDD236 pKa = 3.96SCLSRR241 pKa = 11.84AFNN244 pKa = 3.89
Molecular weight: 26.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2706 |
244 |
1862 |
676.5 |
75.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.948 ± 1.102 | 2.106 ± 0.368 |
6.245 ± 0.498 | 5.617 ± 0.179 |
4.582 ± 0.304 | 5.728 ± 1.155 |
2.402 ± 0.393 | 5.026 ± 0.279 |
5.876 ± 0.932 | 8.684 ± 0.519 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.03 ± 0.34 | 3.732 ± 0.521 |
4.324 ± 0.472 | 2.993 ± 0.284 |
5.58 ± 0.651 | 8.537 ± 0.697 |
5.765 ± 0.318 | 8.389 ± 0.267 |
1.072 ± 0.097 | 3.363 ± 0.435 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
