
Porcine associated porprismacovirus 10
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
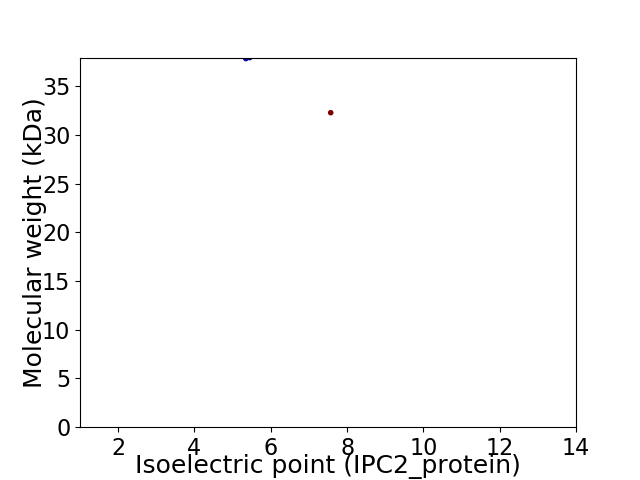
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HWI1|A0A160HWI1_9VIRU Replication associated protein OS=Porcine associated porprismacovirus 10 OX=2170117 PE=4 SV=1
MM1 pKa = 7.18AQSISVSVSEE11 pKa = 4.8TYY13 pKa = 10.77DD14 pKa = 3.3LSTTKK19 pKa = 10.69DD20 pKa = 3.34RR21 pKa = 11.84LGLIAIRR28 pKa = 11.84TPSMIAVNKK37 pKa = 9.67RR38 pKa = 11.84YY39 pKa = 9.49PGFIRR44 pKa = 11.84NFKK47 pKa = 9.18FLKK50 pKa = 10.24VKK52 pKa = 10.51SADD55 pKa = 3.56VVLSCASSLPADD67 pKa = 4.11PLQIGTTAGAIAPQDD82 pKa = 3.48MFNPLLYY89 pKa = 10.4KK90 pKa = 10.52AVSNDD95 pKa = 2.75SWNGLINRR103 pKa = 11.84IYY105 pKa = 10.89AGGLGLSSDD114 pKa = 3.96SLGGSVRR121 pKa = 11.84YY122 pKa = 8.58FQDD125 pKa = 3.75AFPQSSNSDD134 pKa = 3.6CMAMYY139 pKa = 10.62YY140 pKa = 10.62SLLSDD145 pKa = 3.77PSFKK149 pKa = 10.23KK150 pKa = 10.54AHH152 pKa = 5.18VQSGLEE158 pKa = 3.9MRR160 pKa = 11.84HH161 pKa = 6.51LIPLVYY167 pKa = 10.15HH168 pKa = 6.73LLSSGGVVPANNGAQIAGNASMNDD192 pKa = 3.17QVYY195 pKa = 10.41IGSNNIAGTSSILAYY210 pKa = 10.65NGAGSMKK217 pKa = 9.72GRR219 pKa = 11.84PVPMPPVEE227 pKa = 4.37CTPSVYY233 pKa = 10.46HH234 pKa = 5.53EE235 pKa = 4.26TQDD238 pKa = 3.38NVVRR242 pKa = 11.84GEE244 pKa = 3.85WTPTVGNIVPTYY256 pKa = 8.5VGCIIVPPASLNITYY271 pKa = 9.98FRR273 pKa = 11.84MTIRR277 pKa = 11.84WNLEE281 pKa = 3.73FFGVASDD288 pKa = 3.44IAKK291 pKa = 10.34ALPTSAVAISPYY303 pKa = 10.29TYY305 pKa = 10.48FSNQQTQQTLQASKK319 pKa = 10.47ISDD322 pKa = 3.59VSDD325 pKa = 3.27TAVNADD331 pKa = 3.68GDD333 pKa = 4.15DD334 pKa = 3.7GFARR338 pKa = 11.84SVEE341 pKa = 4.07ADD343 pKa = 3.49GVTLDD348 pKa = 5.71LIMEE352 pKa = 4.66HH353 pKa = 6.8
MM1 pKa = 7.18AQSISVSVSEE11 pKa = 4.8TYY13 pKa = 10.77DD14 pKa = 3.3LSTTKK19 pKa = 10.69DD20 pKa = 3.34RR21 pKa = 11.84LGLIAIRR28 pKa = 11.84TPSMIAVNKK37 pKa = 9.67RR38 pKa = 11.84YY39 pKa = 9.49PGFIRR44 pKa = 11.84NFKK47 pKa = 9.18FLKK50 pKa = 10.24VKK52 pKa = 10.51SADD55 pKa = 3.56VVLSCASSLPADD67 pKa = 4.11PLQIGTTAGAIAPQDD82 pKa = 3.48MFNPLLYY89 pKa = 10.4KK90 pKa = 10.52AVSNDD95 pKa = 2.75SWNGLINRR103 pKa = 11.84IYY105 pKa = 10.89AGGLGLSSDD114 pKa = 3.96SLGGSVRR121 pKa = 11.84YY122 pKa = 8.58FQDD125 pKa = 3.75AFPQSSNSDD134 pKa = 3.6CMAMYY139 pKa = 10.62YY140 pKa = 10.62SLLSDD145 pKa = 3.77PSFKK149 pKa = 10.23KK150 pKa = 10.54AHH152 pKa = 5.18VQSGLEE158 pKa = 3.9MRR160 pKa = 11.84HH161 pKa = 6.51LIPLVYY167 pKa = 10.15HH168 pKa = 6.73LLSSGGVVPANNGAQIAGNASMNDD192 pKa = 3.17QVYY195 pKa = 10.41IGSNNIAGTSSILAYY210 pKa = 10.65NGAGSMKK217 pKa = 9.72GRR219 pKa = 11.84PVPMPPVEE227 pKa = 4.37CTPSVYY233 pKa = 10.46HH234 pKa = 5.53EE235 pKa = 4.26TQDD238 pKa = 3.38NVVRR242 pKa = 11.84GEE244 pKa = 3.85WTPTVGNIVPTYY256 pKa = 8.5VGCIIVPPASLNITYY271 pKa = 9.98FRR273 pKa = 11.84MTIRR277 pKa = 11.84WNLEE281 pKa = 3.73FFGVASDD288 pKa = 3.44IAKK291 pKa = 10.34ALPTSAVAISPYY303 pKa = 10.29TYY305 pKa = 10.48FSNQQTQQTLQASKK319 pKa = 10.47ISDD322 pKa = 3.59VSDD325 pKa = 3.27TAVNADD331 pKa = 3.68GDD333 pKa = 4.15DD334 pKa = 3.7GFARR338 pKa = 11.84SVEE341 pKa = 4.07ADD343 pKa = 3.49GVTLDD348 pKa = 5.71LIMEE352 pKa = 4.66HH353 pKa = 6.8
Molecular weight: 37.82 kDa
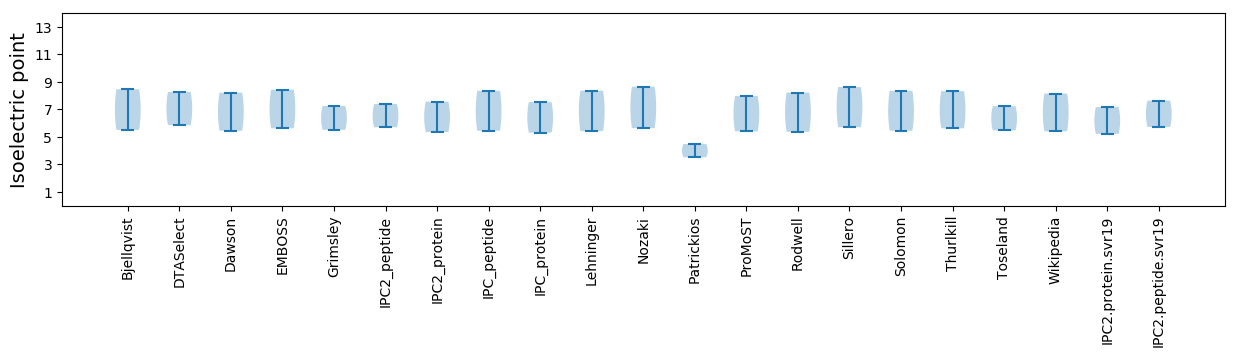
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HWI1|A0A160HWI1_9VIRU Replication associated protein OS=Porcine associated porprismacovirus 10 OX=2170117 PE=4 SV=1
MM1 pKa = 6.16QTWVLTIPRR10 pKa = 11.84RR11 pKa = 11.84TEE13 pKa = 3.5IQVPTWLWLAWEE25 pKa = 4.34PVTIVSVCSKK35 pKa = 10.93KK36 pKa = 10.49LLNYY40 pKa = 9.89ILEE43 pKa = 4.21QSDD46 pKa = 3.42TKK48 pKa = 10.8RR49 pKa = 11.84YY50 pKa = 9.75IIGIEE55 pKa = 3.76KK56 pKa = 10.04GKK58 pKa = 10.54NGLEE62 pKa = 3.93HH63 pKa = 5.73FQIRR67 pKa = 11.84LSCSDD72 pKa = 3.68PEE74 pKa = 4.45FFEE77 pKa = 5.86HH78 pKa = 5.72MKK80 pKa = 9.98EE81 pKa = 3.88WYY83 pKa = 9.08PYY85 pKa = 10.28AHH87 pKa = 6.99IEE89 pKa = 4.22KK90 pKa = 9.97SDD92 pKa = 3.32VGINSEE98 pKa = 3.97SMEE101 pKa = 4.09YY102 pKa = 9.83EE103 pKa = 4.09RR104 pKa = 11.84KK105 pKa = 9.65EE106 pKa = 3.85GRR108 pKa = 11.84YY109 pKa = 4.94WTSMDD114 pKa = 3.08TTEE117 pKa = 5.26IRR119 pKa = 11.84IQRR122 pKa = 11.84FGKK125 pKa = 9.56PNRR128 pKa = 11.84TQQRR132 pKa = 11.84VLEE135 pKa = 4.16VLRR138 pKa = 11.84RR139 pKa = 11.84TNDD142 pKa = 3.12RR143 pKa = 11.84EE144 pKa = 3.99IVLWYY149 pKa = 9.88SDD151 pKa = 3.48KK152 pKa = 11.5GSIGKK157 pKa = 8.77SWLVGHH163 pKa = 6.85LWEE166 pKa = 4.45TGEE169 pKa = 5.42AYY171 pKa = 10.34VCQPQDD177 pKa = 3.08TVKK180 pKa = 10.81GMKK183 pKa = 9.55QDD185 pKa = 3.38VASEE189 pKa = 4.35YY190 pKa = 10.04IKK192 pKa = 10.85HH193 pKa = 5.78GWRR196 pKa = 11.84PCIVVDD202 pKa = 6.37LPRR205 pKa = 11.84TWKK208 pKa = 7.21WTKK211 pKa = 10.75DD212 pKa = 3.26LYY214 pKa = 11.06CALEE218 pKa = 4.27SIKK221 pKa = 11.02DD222 pKa = 3.74GLLKK226 pKa = 9.85DD227 pKa = 3.64TRR229 pKa = 11.84YY230 pKa = 10.8SSDD233 pKa = 3.33TINIKK238 pKa = 9.35GVKK241 pKa = 9.88VLVTSNTLPKK251 pKa = 9.98FDD253 pKa = 4.49SLSFDD258 pKa = 2.69RR259 pKa = 11.84WIVIEE264 pKa = 3.92NTIEE268 pKa = 3.83RR269 pKa = 11.84RR270 pKa = 11.84SFSS273 pKa = 3.28
MM1 pKa = 6.16QTWVLTIPRR10 pKa = 11.84RR11 pKa = 11.84TEE13 pKa = 3.5IQVPTWLWLAWEE25 pKa = 4.34PVTIVSVCSKK35 pKa = 10.93KK36 pKa = 10.49LLNYY40 pKa = 9.89ILEE43 pKa = 4.21QSDD46 pKa = 3.42TKK48 pKa = 10.8RR49 pKa = 11.84YY50 pKa = 9.75IIGIEE55 pKa = 3.76KK56 pKa = 10.04GKK58 pKa = 10.54NGLEE62 pKa = 3.93HH63 pKa = 5.73FQIRR67 pKa = 11.84LSCSDD72 pKa = 3.68PEE74 pKa = 4.45FFEE77 pKa = 5.86HH78 pKa = 5.72MKK80 pKa = 9.98EE81 pKa = 3.88WYY83 pKa = 9.08PYY85 pKa = 10.28AHH87 pKa = 6.99IEE89 pKa = 4.22KK90 pKa = 9.97SDD92 pKa = 3.32VGINSEE98 pKa = 3.97SMEE101 pKa = 4.09YY102 pKa = 9.83EE103 pKa = 4.09RR104 pKa = 11.84KK105 pKa = 9.65EE106 pKa = 3.85GRR108 pKa = 11.84YY109 pKa = 4.94WTSMDD114 pKa = 3.08TTEE117 pKa = 5.26IRR119 pKa = 11.84IQRR122 pKa = 11.84FGKK125 pKa = 9.56PNRR128 pKa = 11.84TQQRR132 pKa = 11.84VLEE135 pKa = 4.16VLRR138 pKa = 11.84RR139 pKa = 11.84TNDD142 pKa = 3.12RR143 pKa = 11.84EE144 pKa = 3.99IVLWYY149 pKa = 9.88SDD151 pKa = 3.48KK152 pKa = 11.5GSIGKK157 pKa = 8.77SWLVGHH163 pKa = 6.85LWEE166 pKa = 4.45TGEE169 pKa = 5.42AYY171 pKa = 10.34VCQPQDD177 pKa = 3.08TVKK180 pKa = 10.81GMKK183 pKa = 9.55QDD185 pKa = 3.38VASEE189 pKa = 4.35YY190 pKa = 10.04IKK192 pKa = 10.85HH193 pKa = 5.78GWRR196 pKa = 11.84PCIVVDD202 pKa = 6.37LPRR205 pKa = 11.84TWKK208 pKa = 7.21WTKK211 pKa = 10.75DD212 pKa = 3.26LYY214 pKa = 11.06CALEE218 pKa = 4.27SIKK221 pKa = 11.02DD222 pKa = 3.74GLLKK226 pKa = 9.85DD227 pKa = 3.64TRR229 pKa = 11.84YY230 pKa = 10.8SSDD233 pKa = 3.33TINIKK238 pKa = 9.35GVKK241 pKa = 9.88VLVTSNTLPKK251 pKa = 9.98FDD253 pKa = 4.49SLSFDD258 pKa = 2.69RR259 pKa = 11.84WIVIEE264 pKa = 3.92NTIEE268 pKa = 3.83RR269 pKa = 11.84RR270 pKa = 11.84SFSS273 pKa = 3.28
Molecular weight: 32.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
626 |
273 |
353 |
313.0 |
35.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.911 ± 2.446 | 1.438 ± 0.236 |
5.591 ± 0.058 | 4.792 ± 1.959 |
3.035 ± 0.282 | 6.55 ± 0.852 |
1.597 ± 0.14 | 7.188 ± 0.522 |
5.112 ± 1.547 | 7.668 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.556 ± 0.434 | 4.473 ± 0.925 |
4.952 ± 0.773 | 3.834 ± 0.102 |
4.952 ± 1.204 | 9.744 ± 1.231 |
6.39 ± 0.561 | 7.508 ± 0.329 |
2.556 ± 1.323 | 4.153 ± 0.074 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
