
Rodent associated circovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus
Average proteome isoelectric point is 7.76
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4MWX5|A0A2H4MWX5_9CIRC ATP-dependent helicase Rep OS=Rodent associated circovirus 1 OX=2560708 PE=3 SV=1
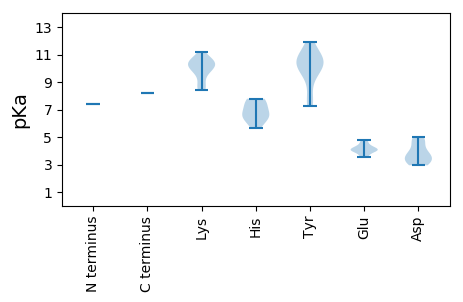
MM1 pKa = 7.4AQAQILQQRR10 pKa = 11.84LVKK13 pKa = 10.48QGARR17 pKa = 11.84GRR19 pKa = 11.84WCFTINNYY27 pKa = 7.26TAEE30 pKa = 4.14EE31 pKa = 4.02EE32 pKa = 4.22QAVIALAGEE41 pKa = 4.34AKK43 pKa = 10.6YY44 pKa = 10.04LICGRR49 pKa = 11.84EE50 pKa = 3.93VGEE53 pKa = 4.53NGTPHH58 pKa = 6.46LQGFVNLKK66 pKa = 8.66KK67 pKa = 10.33KK68 pKa = 10.48KK69 pKa = 9.97RR70 pKa = 11.84FAAMKK75 pKa = 8.46EE76 pKa = 3.96ALGGRR81 pKa = 11.84CHH83 pKa = 7.71LEE85 pKa = 3.51PARR88 pKa = 11.84GDD90 pKa = 3.43DD91 pKa = 4.68CSNKK95 pKa = 10.16DD96 pKa = 3.56YY97 pKa = 10.78CSKK100 pKa = 11.16GGDD103 pKa = 3.21ILIEE107 pKa = 4.02EE108 pKa = 4.72GVPQRR113 pKa = 11.84EE114 pKa = 4.16RR115 pKa = 11.84QRR117 pKa = 11.84TDD119 pKa = 2.95LKK121 pKa = 10.64QACEE125 pKa = 4.05LVTQGGDD132 pKa = 2.95IRR134 pKa = 11.84HH135 pKa = 5.68VARR138 pKa = 11.84DD139 pKa = 3.61YY140 pKa = 11.1PEE142 pKa = 4.1VFVRR146 pKa = 11.84YY147 pKa = 9.36HH148 pKa = 6.75RR149 pKa = 11.84GLLALQLYY157 pKa = 10.01HH158 pKa = 7.78PDD160 pKa = 3.2LCLPRR165 pKa = 11.84QWKK168 pKa = 8.43TEE170 pKa = 3.65VFVYY174 pKa = 10.19VGPPGVGKK182 pKa = 10.62SRR184 pKa = 11.84IVQAKK189 pKa = 10.17APIGYY194 pKa = 7.81WKK196 pKa = 10.03PRR198 pKa = 11.84GKK200 pKa = 9.03WWDD203 pKa = 3.98GYY205 pKa = 10.81HH206 pKa = 6.46GNSDD210 pKa = 3.91VILDD214 pKa = 4.7DD215 pKa = 4.21FYY217 pKa = 11.95GWLPFDD223 pKa = 5.03DD224 pKa = 4.89LLRR227 pKa = 11.84MCDD230 pKa = 3.54RR231 pKa = 11.84YY232 pKa = 10.52PLTVEE237 pKa = 4.33TKK239 pKa = 10.58GGTTQFLAKK248 pKa = 10.19RR249 pKa = 11.84IFITSNKK256 pKa = 9.5SPSEE260 pKa = 4.11WYY262 pKa = 10.28SDD264 pKa = 3.6EE265 pKa = 3.98ITNKK269 pKa = 9.76DD270 pKa = 3.09ALYY273 pKa = 10.56RR274 pKa = 11.84RR275 pKa = 11.84LTEE278 pKa = 3.61LHH280 pKa = 6.29MWTGTCFEE288 pKa = 4.75HH289 pKa = 7.1PPPRR293 pKa = 11.84VMWNHH298 pKa = 7.32KK299 pKa = 9.92INYY302 pKa = 8.23
MM1 pKa = 7.4AQAQILQQRR10 pKa = 11.84LVKK13 pKa = 10.48QGARR17 pKa = 11.84GRR19 pKa = 11.84WCFTINNYY27 pKa = 7.26TAEE30 pKa = 4.14EE31 pKa = 4.02EE32 pKa = 4.22QAVIALAGEE41 pKa = 4.34AKK43 pKa = 10.6YY44 pKa = 10.04LICGRR49 pKa = 11.84EE50 pKa = 3.93VGEE53 pKa = 4.53NGTPHH58 pKa = 6.46LQGFVNLKK66 pKa = 8.66KK67 pKa = 10.33KK68 pKa = 10.48KK69 pKa = 9.97RR70 pKa = 11.84FAAMKK75 pKa = 8.46EE76 pKa = 3.96ALGGRR81 pKa = 11.84CHH83 pKa = 7.71LEE85 pKa = 3.51PARR88 pKa = 11.84GDD90 pKa = 3.43DD91 pKa = 4.68CSNKK95 pKa = 10.16DD96 pKa = 3.56YY97 pKa = 10.78CSKK100 pKa = 11.16GGDD103 pKa = 3.21ILIEE107 pKa = 4.02EE108 pKa = 4.72GVPQRR113 pKa = 11.84EE114 pKa = 4.16RR115 pKa = 11.84QRR117 pKa = 11.84TDD119 pKa = 2.95LKK121 pKa = 10.64QACEE125 pKa = 4.05LVTQGGDD132 pKa = 2.95IRR134 pKa = 11.84HH135 pKa = 5.68VARR138 pKa = 11.84DD139 pKa = 3.61YY140 pKa = 11.1PEE142 pKa = 4.1VFVRR146 pKa = 11.84YY147 pKa = 9.36HH148 pKa = 6.75RR149 pKa = 11.84GLLALQLYY157 pKa = 10.01HH158 pKa = 7.78PDD160 pKa = 3.2LCLPRR165 pKa = 11.84QWKK168 pKa = 8.43TEE170 pKa = 3.65VFVYY174 pKa = 10.19VGPPGVGKK182 pKa = 10.62SRR184 pKa = 11.84IVQAKK189 pKa = 10.17APIGYY194 pKa = 7.81WKK196 pKa = 10.03PRR198 pKa = 11.84GKK200 pKa = 9.03WWDD203 pKa = 3.98GYY205 pKa = 10.81HH206 pKa = 6.46GNSDD210 pKa = 3.91VILDD214 pKa = 4.7DD215 pKa = 4.21FYY217 pKa = 11.95GWLPFDD223 pKa = 5.03DD224 pKa = 4.89LLRR227 pKa = 11.84MCDD230 pKa = 3.54RR231 pKa = 11.84YY232 pKa = 10.52PLTVEE237 pKa = 4.33TKK239 pKa = 10.58GGTTQFLAKK248 pKa = 10.19RR249 pKa = 11.84IFITSNKK256 pKa = 9.5SPSEE260 pKa = 4.11WYY262 pKa = 10.28SDD264 pKa = 3.6EE265 pKa = 3.98ITNKK269 pKa = 9.76DD270 pKa = 3.09ALYY273 pKa = 10.56RR274 pKa = 11.84RR275 pKa = 11.84LTEE278 pKa = 3.61LHH280 pKa = 6.29MWTGTCFEE288 pKa = 4.75HH289 pKa = 7.1PPPRR293 pKa = 11.84VMWNHH298 pKa = 7.32KK299 pKa = 9.92INYY302 pKa = 8.23
Molecular weight: 34.77 kDa
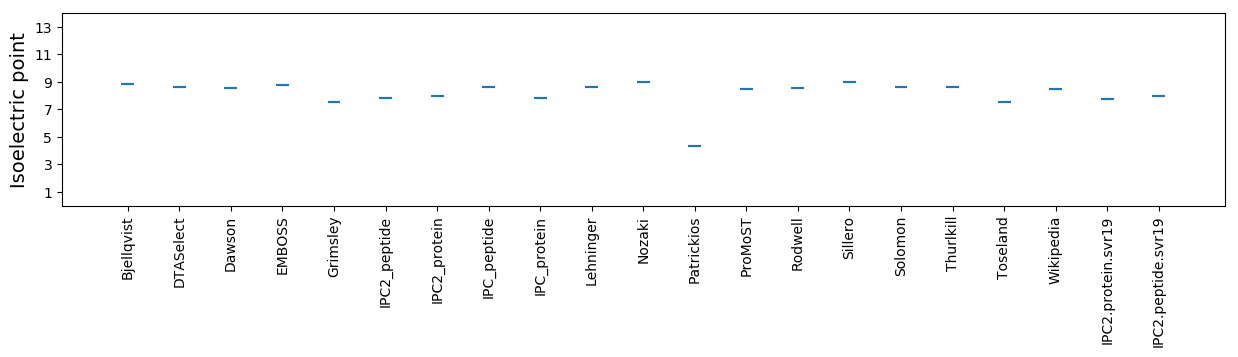
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4MWX5|A0A2H4MWX5_9CIRC ATP-dependent helicase Rep OS=Rodent associated circovirus 1 OX=2560708 PE=3 SV=1
MM1 pKa = 7.4AQAQILQQRR10 pKa = 11.84LVKK13 pKa = 10.48QGARR17 pKa = 11.84GRR19 pKa = 11.84WCFTINNYY27 pKa = 7.26TAEE30 pKa = 4.14EE31 pKa = 4.02EE32 pKa = 4.22QAVIALAGEE41 pKa = 4.34AKK43 pKa = 10.6YY44 pKa = 10.04LICGRR49 pKa = 11.84EE50 pKa = 3.93VGEE53 pKa = 4.53NGTPHH58 pKa = 6.46LQGFVNLKK66 pKa = 8.66KK67 pKa = 10.33KK68 pKa = 10.48KK69 pKa = 9.97RR70 pKa = 11.84FAAMKK75 pKa = 8.46EE76 pKa = 3.96ALGGRR81 pKa = 11.84CHH83 pKa = 7.71LEE85 pKa = 3.51PARR88 pKa = 11.84GDD90 pKa = 3.43DD91 pKa = 4.68CSNKK95 pKa = 10.16DD96 pKa = 3.56YY97 pKa = 10.78CSKK100 pKa = 11.16GGDD103 pKa = 3.21ILIEE107 pKa = 4.02EE108 pKa = 4.72GVPQRR113 pKa = 11.84EE114 pKa = 4.16RR115 pKa = 11.84QRR117 pKa = 11.84TDD119 pKa = 2.95LKK121 pKa = 10.64QACEE125 pKa = 4.05LVTQGGDD132 pKa = 2.95IRR134 pKa = 11.84HH135 pKa = 5.68VARR138 pKa = 11.84DD139 pKa = 3.61YY140 pKa = 11.1PEE142 pKa = 4.1VFVRR146 pKa = 11.84YY147 pKa = 9.36HH148 pKa = 6.75RR149 pKa = 11.84GLLALQLYY157 pKa = 10.01HH158 pKa = 7.78PDD160 pKa = 3.2LCLPRR165 pKa = 11.84QWKK168 pKa = 8.43TEE170 pKa = 3.65VFVYY174 pKa = 10.19VGPPGVGKK182 pKa = 10.62SRR184 pKa = 11.84IVQAKK189 pKa = 10.17APIGYY194 pKa = 7.81WKK196 pKa = 10.03PRR198 pKa = 11.84GKK200 pKa = 9.03WWDD203 pKa = 3.98GYY205 pKa = 10.81HH206 pKa = 6.46GNSDD210 pKa = 3.91VILDD214 pKa = 4.7DD215 pKa = 4.21FYY217 pKa = 11.95GWLPFDD223 pKa = 5.03DD224 pKa = 4.89LLRR227 pKa = 11.84MCDD230 pKa = 3.54RR231 pKa = 11.84YY232 pKa = 10.52PLTVEE237 pKa = 4.33TKK239 pKa = 10.58GGTTQFLAKK248 pKa = 10.19RR249 pKa = 11.84IFITSNKK256 pKa = 9.5SPSEE260 pKa = 4.11WYY262 pKa = 10.28SDD264 pKa = 3.6EE265 pKa = 3.98ITNKK269 pKa = 9.76DD270 pKa = 3.09ALYY273 pKa = 10.56RR274 pKa = 11.84RR275 pKa = 11.84LTEE278 pKa = 3.61LHH280 pKa = 6.29MWTGTCFEE288 pKa = 4.75HH289 pKa = 7.1PPPRR293 pKa = 11.84VMWNHH298 pKa = 7.32KK299 pKa = 9.92INYY302 pKa = 8.23
MM1 pKa = 7.4AQAQILQQRR10 pKa = 11.84LVKK13 pKa = 10.48QGARR17 pKa = 11.84GRR19 pKa = 11.84WCFTINNYY27 pKa = 7.26TAEE30 pKa = 4.14EE31 pKa = 4.02EE32 pKa = 4.22QAVIALAGEE41 pKa = 4.34AKK43 pKa = 10.6YY44 pKa = 10.04LICGRR49 pKa = 11.84EE50 pKa = 3.93VGEE53 pKa = 4.53NGTPHH58 pKa = 6.46LQGFVNLKK66 pKa = 8.66KK67 pKa = 10.33KK68 pKa = 10.48KK69 pKa = 9.97RR70 pKa = 11.84FAAMKK75 pKa = 8.46EE76 pKa = 3.96ALGGRR81 pKa = 11.84CHH83 pKa = 7.71LEE85 pKa = 3.51PARR88 pKa = 11.84GDD90 pKa = 3.43DD91 pKa = 4.68CSNKK95 pKa = 10.16DD96 pKa = 3.56YY97 pKa = 10.78CSKK100 pKa = 11.16GGDD103 pKa = 3.21ILIEE107 pKa = 4.02EE108 pKa = 4.72GVPQRR113 pKa = 11.84EE114 pKa = 4.16RR115 pKa = 11.84QRR117 pKa = 11.84TDD119 pKa = 2.95LKK121 pKa = 10.64QACEE125 pKa = 4.05LVTQGGDD132 pKa = 2.95IRR134 pKa = 11.84HH135 pKa = 5.68VARR138 pKa = 11.84DD139 pKa = 3.61YY140 pKa = 11.1PEE142 pKa = 4.1VFVRR146 pKa = 11.84YY147 pKa = 9.36HH148 pKa = 6.75RR149 pKa = 11.84GLLALQLYY157 pKa = 10.01HH158 pKa = 7.78PDD160 pKa = 3.2LCLPRR165 pKa = 11.84QWKK168 pKa = 8.43TEE170 pKa = 3.65VFVYY174 pKa = 10.19VGPPGVGKK182 pKa = 10.62SRR184 pKa = 11.84IVQAKK189 pKa = 10.17APIGYY194 pKa = 7.81WKK196 pKa = 10.03PRR198 pKa = 11.84GKK200 pKa = 9.03WWDD203 pKa = 3.98GYY205 pKa = 10.81HH206 pKa = 6.46GNSDD210 pKa = 3.91VILDD214 pKa = 4.7DD215 pKa = 4.21FYY217 pKa = 11.95GWLPFDD223 pKa = 5.03DD224 pKa = 4.89LLRR227 pKa = 11.84MCDD230 pKa = 3.54RR231 pKa = 11.84YY232 pKa = 10.52PLTVEE237 pKa = 4.33TKK239 pKa = 10.58GGTTQFLAKK248 pKa = 10.19RR249 pKa = 11.84IFITSNKK256 pKa = 9.5SPSEE260 pKa = 4.11WYY262 pKa = 10.28SDD264 pKa = 3.6EE265 pKa = 3.98ITNKK269 pKa = 9.76DD270 pKa = 3.09ALYY273 pKa = 10.56RR274 pKa = 11.84RR275 pKa = 11.84LTEE278 pKa = 3.61LHH280 pKa = 6.29MWTGTCFEE288 pKa = 4.75HH289 pKa = 7.1PPPRR293 pKa = 11.84VMWNHH298 pKa = 7.32KK299 pKa = 9.92INYY302 pKa = 8.23
Molecular weight: 34.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
302 |
302 |
302 |
302.0 |
34.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.291 ± 0.0 | 2.98 ± 0.0 |
5.629 ± 0.0 | 6.291 ± 0.0 |
3.311 ± 0.0 | 8.94 ± 0.0 |
2.98 ± 0.0 | 4.636 ± 0.0 |
6.623 ± 0.0 | 8.609 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.656 ± 0.0 | 3.311 ± 0.0 |
5.298 ± 0.0 | 4.967 ± 0.0 |
7.616 ± 0.0 | 2.649 ± 0.0 |
4.967 ± 0.0 | 5.629 ± 0.0 |
2.98 ± 0.0 | 4.636 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
