
Heterosigma akashiwo virus 01 (HaV01)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Algavirales; Phycodnaviridae; Raphidovirus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
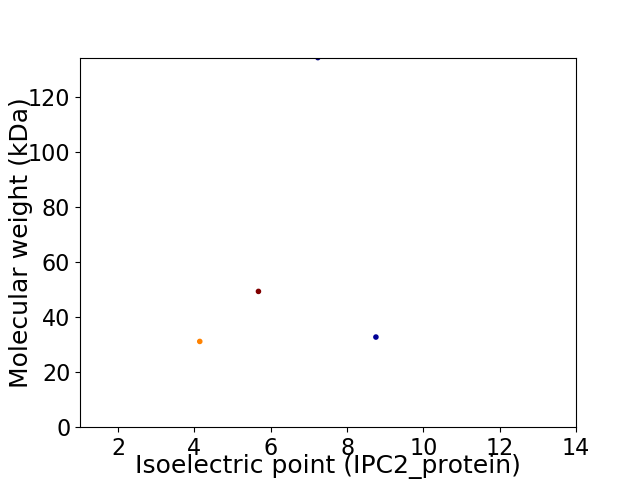
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91DI1|Q91DI1_HAV01 UKCH-2 OS=Heterosigma akashiwo virus 01 OX=97195 GN=ukch-2 PE=4 SV=1
MM1 pKa = 7.28FVRR4 pKa = 11.84IVFLKK9 pKa = 10.69SLLILYY15 pKa = 10.15SNILMDD21 pKa = 4.5SLDD24 pKa = 3.63IRR26 pKa = 11.84DD27 pKa = 3.87PSNFTFTEE35 pKa = 4.45DD36 pKa = 3.28EE37 pKa = 4.09QKK39 pKa = 11.11EE40 pKa = 3.97IDD42 pKa = 3.76KK43 pKa = 10.66KK44 pKa = 10.9LNEE47 pKa = 5.28AIPEE51 pKa = 4.14LSDD54 pKa = 3.57KK55 pKa = 10.88NVNLEE60 pKa = 3.89TDD62 pKa = 3.68HH63 pKa = 7.9IKK65 pKa = 10.26IDD67 pKa = 3.78GQNFALISIVAPEE80 pKa = 4.13GLNQKK85 pKa = 8.76SNKK88 pKa = 9.29VCIKK92 pKa = 10.05IKK94 pKa = 10.69GVFDD98 pKa = 3.54TVEE101 pKa = 3.95NARR104 pKa = 11.84QHH106 pKa = 6.83ANDD109 pKa = 4.07LTKK112 pKa = 10.31HH113 pKa = 6.57DD114 pKa = 3.82PTFDD118 pKa = 3.5VFVVSLYY125 pKa = 10.48EE126 pKa = 3.95WLLVPPDD133 pKa = 3.45MDD135 pKa = 4.74KK136 pKa = 10.78ISEE139 pKa = 4.0QEE141 pKa = 4.2YY142 pKa = 10.9VDD144 pKa = 4.67EE145 pKa = 4.59EE146 pKa = 4.89LNTIITEE153 pKa = 4.48FKK155 pKa = 10.47KK156 pKa = 10.59SQKK159 pKa = 9.39QVQLEE164 pKa = 4.05FEE166 pKa = 4.3TRR168 pKa = 11.84KK169 pKa = 10.31DD170 pKa = 3.43ALRR173 pKa = 11.84SNIDD177 pKa = 2.99INTTIINDD185 pKa = 2.95EE186 pKa = 4.16DD187 pKa = 4.19MNVKK191 pKa = 9.14MLPNEE196 pKa = 4.05EE197 pKa = 5.12LGDD200 pKa = 4.16DD201 pKa = 3.6SDD203 pKa = 5.04GGVEE207 pKa = 3.89EE208 pKa = 4.56TKK210 pKa = 10.98GEE212 pKa = 4.07KK213 pKa = 9.78HH214 pKa = 6.02VSFSNDD220 pKa = 2.28IGTLLEE226 pKa = 4.59KK227 pKa = 10.66MNGTDD232 pKa = 4.57IGTLLEE238 pKa = 4.58KK239 pKa = 10.66MNGTDD244 pKa = 3.42SWCKK248 pKa = 10.96DD249 pKa = 3.43EE250 pKa = 7.32DD251 pKa = 5.87DD252 pKa = 6.3DD253 pKa = 7.56DD254 pKa = 6.74DD255 pKa = 7.23DD256 pKa = 6.55NDD258 pKa = 4.8DD259 pKa = 5.08DD260 pKa = 6.87DD261 pKa = 6.73DD262 pKa = 4.77IKK264 pKa = 11.15EE265 pKa = 4.46LKK267 pKa = 10.24NDD269 pKa = 3.75LKK271 pKa = 10.84IVV273 pKa = 3.47
MM1 pKa = 7.28FVRR4 pKa = 11.84IVFLKK9 pKa = 10.69SLLILYY15 pKa = 10.15SNILMDD21 pKa = 4.5SLDD24 pKa = 3.63IRR26 pKa = 11.84DD27 pKa = 3.87PSNFTFTEE35 pKa = 4.45DD36 pKa = 3.28EE37 pKa = 4.09QKK39 pKa = 11.11EE40 pKa = 3.97IDD42 pKa = 3.76KK43 pKa = 10.66KK44 pKa = 10.9LNEE47 pKa = 5.28AIPEE51 pKa = 4.14LSDD54 pKa = 3.57KK55 pKa = 10.88NVNLEE60 pKa = 3.89TDD62 pKa = 3.68HH63 pKa = 7.9IKK65 pKa = 10.26IDD67 pKa = 3.78GQNFALISIVAPEE80 pKa = 4.13GLNQKK85 pKa = 8.76SNKK88 pKa = 9.29VCIKK92 pKa = 10.05IKK94 pKa = 10.69GVFDD98 pKa = 3.54TVEE101 pKa = 3.95NARR104 pKa = 11.84QHH106 pKa = 6.83ANDD109 pKa = 4.07LTKK112 pKa = 10.31HH113 pKa = 6.57DD114 pKa = 3.82PTFDD118 pKa = 3.5VFVVSLYY125 pKa = 10.48EE126 pKa = 3.95WLLVPPDD133 pKa = 3.45MDD135 pKa = 4.74KK136 pKa = 10.78ISEE139 pKa = 4.0QEE141 pKa = 4.2YY142 pKa = 10.9VDD144 pKa = 4.67EE145 pKa = 4.59EE146 pKa = 4.89LNTIITEE153 pKa = 4.48FKK155 pKa = 10.47KK156 pKa = 10.59SQKK159 pKa = 9.39QVQLEE164 pKa = 4.05FEE166 pKa = 4.3TRR168 pKa = 11.84KK169 pKa = 10.31DD170 pKa = 3.43ALRR173 pKa = 11.84SNIDD177 pKa = 2.99INTTIINDD185 pKa = 2.95EE186 pKa = 4.16DD187 pKa = 4.19MNVKK191 pKa = 9.14MLPNEE196 pKa = 4.05EE197 pKa = 5.12LGDD200 pKa = 4.16DD201 pKa = 3.6SDD203 pKa = 5.04GGVEE207 pKa = 3.89EE208 pKa = 4.56TKK210 pKa = 10.98GEE212 pKa = 4.07KK213 pKa = 9.78HH214 pKa = 6.02VSFSNDD220 pKa = 2.28IGTLLEE226 pKa = 4.59KK227 pKa = 10.66MNGTDD232 pKa = 4.57IGTLLEE238 pKa = 4.58KK239 pKa = 10.66MNGTDD244 pKa = 3.42SWCKK248 pKa = 10.96DD249 pKa = 3.43EE250 pKa = 7.32DD251 pKa = 5.87DD252 pKa = 6.3DD253 pKa = 7.56DD254 pKa = 6.74DD255 pKa = 7.23DD256 pKa = 6.55NDD258 pKa = 4.8DD259 pKa = 5.08DD260 pKa = 6.87DD261 pKa = 6.73DD262 pKa = 4.77IKK264 pKa = 11.15EE265 pKa = 4.46LKK267 pKa = 10.24NDD269 pKa = 3.75LKK271 pKa = 10.84IVV273 pKa = 3.47
Molecular weight: 31.2 kDa
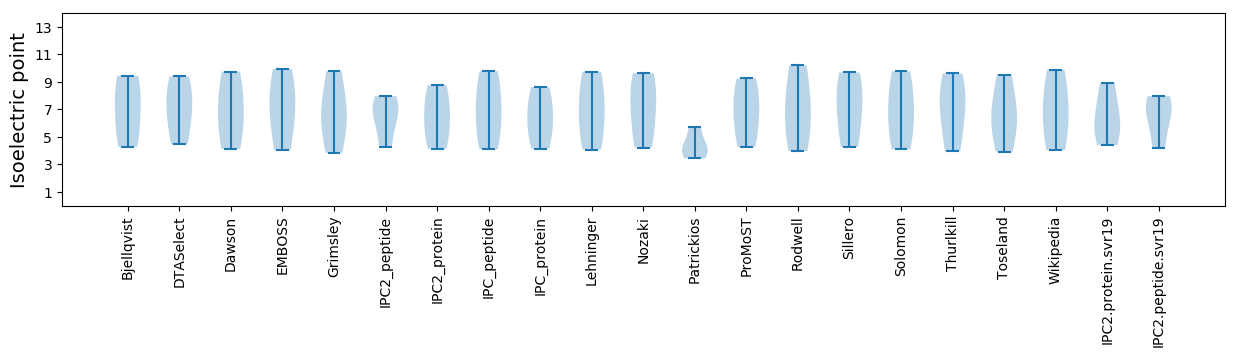
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91DI1|Q91DI1_HAV01 UKCH-2 OS=Heterosigma akashiwo virus 01 OX=97195 GN=ukch-2 PE=4 SV=1
MM1 pKa = 7.65EE2 pKa = 4.57LQIKK6 pKa = 10.22KK7 pKa = 10.03FDD9 pKa = 3.54NRR11 pKa = 11.84SVVDD15 pKa = 3.4KK16 pKa = 10.51RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.5FSSPIIVVIGKK30 pKa = 9.88RR31 pKa = 11.84NTGKK35 pKa = 10.22SEE37 pKa = 4.37LIKK40 pKa = 11.02NIMFCNSDD48 pKa = 3.41IPSGIIISPTEE59 pKa = 4.07SGNSFYY65 pKa = 11.29SDD67 pKa = 3.92FCPGIFVHH75 pKa = 6.02SQYY78 pKa = 11.36DD79 pKa = 3.39SEE81 pKa = 4.84LIKK84 pKa = 10.71KK85 pKa = 9.75IIKK88 pKa = 9.35RR89 pKa = 11.84QKK91 pKa = 10.79KK92 pKa = 9.6LIKK95 pKa = 10.25KK96 pKa = 9.35KK97 pKa = 10.59GKK99 pKa = 9.66HH100 pKa = 5.61PSNDD104 pKa = 2.84FFIILDD110 pKa = 3.63DD111 pKa = 4.15CMYY114 pKa = 10.79DD115 pKa = 2.92SKK117 pKa = 10.32TIGRR121 pKa = 11.84DD122 pKa = 2.9INIRR126 pKa = 11.84EE127 pKa = 3.74IFLNGRR133 pKa = 11.84HH134 pKa = 4.86YY135 pKa = 10.86QIFLILSLQYY145 pKa = 11.66VMDD148 pKa = 4.63LPVSLRR154 pKa = 11.84SNIDD158 pKa = 3.04YY159 pKa = 9.52VFCLRR164 pKa = 11.84EE165 pKa = 3.86NNLQNVEE172 pKa = 4.15KK173 pKa = 10.26LYY175 pKa = 11.15KK176 pKa = 10.21SFFGIFPSKK185 pKa = 10.5IFFNQAFSKK194 pKa = 9.22ITEE197 pKa = 4.21NYY199 pKa = 9.87GSIVLDD205 pKa = 3.16NTSRR209 pKa = 11.84SNRR212 pKa = 11.84IDD214 pKa = 3.24EE215 pKa = 4.63CIFWFRR221 pKa = 11.84ATFPLPNFRR230 pKa = 11.84IGNKK234 pKa = 9.74KK235 pKa = 8.38LWKK238 pKa = 9.14MHH240 pKa = 6.23DD241 pKa = 3.32KK242 pKa = 10.92YY243 pKa = 11.57YY244 pKa = 10.9EE245 pKa = 4.37DD246 pKa = 4.11VSDD249 pKa = 3.88ASEE252 pKa = 4.4DD253 pKa = 3.75EE254 pKa = 4.56KK255 pKa = 11.23EE256 pKa = 4.36SKK258 pKa = 10.97AFDD261 pKa = 4.75DD262 pKa = 4.33IKK264 pKa = 10.42TKK266 pKa = 10.3TSQKK270 pKa = 10.71LKK272 pKa = 10.08TILTNKK278 pKa = 8.38TKK280 pKa = 10.88
MM1 pKa = 7.65EE2 pKa = 4.57LQIKK6 pKa = 10.22KK7 pKa = 10.03FDD9 pKa = 3.54NRR11 pKa = 11.84SVVDD15 pKa = 3.4KK16 pKa = 10.51RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.5FSSPIIVVIGKK30 pKa = 9.88RR31 pKa = 11.84NTGKK35 pKa = 10.22SEE37 pKa = 4.37LIKK40 pKa = 11.02NIMFCNSDD48 pKa = 3.41IPSGIIISPTEE59 pKa = 4.07SGNSFYY65 pKa = 11.29SDD67 pKa = 3.92FCPGIFVHH75 pKa = 6.02SQYY78 pKa = 11.36DD79 pKa = 3.39SEE81 pKa = 4.84LIKK84 pKa = 10.71KK85 pKa = 9.75IIKK88 pKa = 9.35RR89 pKa = 11.84QKK91 pKa = 10.79KK92 pKa = 9.6LIKK95 pKa = 10.25KK96 pKa = 9.35KK97 pKa = 10.59GKK99 pKa = 9.66HH100 pKa = 5.61PSNDD104 pKa = 2.84FFIILDD110 pKa = 3.63DD111 pKa = 4.15CMYY114 pKa = 10.79DD115 pKa = 2.92SKK117 pKa = 10.32TIGRR121 pKa = 11.84DD122 pKa = 2.9INIRR126 pKa = 11.84EE127 pKa = 3.74IFLNGRR133 pKa = 11.84HH134 pKa = 4.86YY135 pKa = 10.86QIFLILSLQYY145 pKa = 11.66VMDD148 pKa = 4.63LPVSLRR154 pKa = 11.84SNIDD158 pKa = 3.04YY159 pKa = 9.52VFCLRR164 pKa = 11.84EE165 pKa = 3.86NNLQNVEE172 pKa = 4.15KK173 pKa = 10.26LYY175 pKa = 11.15KK176 pKa = 10.21SFFGIFPSKK185 pKa = 10.5IFFNQAFSKK194 pKa = 9.22ITEE197 pKa = 4.21NYY199 pKa = 9.87GSIVLDD205 pKa = 3.16NTSRR209 pKa = 11.84SNRR212 pKa = 11.84IDD214 pKa = 3.24EE215 pKa = 4.63CIFWFRR221 pKa = 11.84ATFPLPNFRR230 pKa = 11.84IGNKK234 pKa = 9.74KK235 pKa = 8.38LWKK238 pKa = 9.14MHH240 pKa = 6.23DD241 pKa = 3.32KK242 pKa = 10.92YY243 pKa = 11.57YY244 pKa = 10.9EE245 pKa = 4.37DD246 pKa = 4.11VSDD249 pKa = 3.88ASEE252 pKa = 4.4DD253 pKa = 3.75EE254 pKa = 4.56KK255 pKa = 11.23EE256 pKa = 4.36SKK258 pKa = 10.97AFDD261 pKa = 4.75DD262 pKa = 4.33IKK264 pKa = 10.42TKK266 pKa = 10.3TSQKK270 pKa = 10.71LKK272 pKa = 10.08TILTNKK278 pKa = 8.38TKK280 pKa = 10.88
Molecular weight: 32.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2137 |
273 |
1144 |
534.3 |
61.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.463 ± 1.057 | 1.451 ± 0.253 |
6.27 ± 1.702 | 7.534 ± 1.061 |
5.335 ± 0.509 | 4.82 ± 0.68 |
1.919 ± 0.288 | 8.985 ± 0.825 |
8.142 ± 1.464 | 8.049 ± 0.659 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.854 ± 0.624 | 6.785 ± 0.277 |
3.369 ± 0.184 | 3.369 ± 0.523 |
4.633 ± 0.635 | 6.036 ± 0.744 |
6.177 ± 0.72 | 5.615 ± 0.354 |
1.029 ± 0.113 | 4.165 ± 0.706 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
