
Alternaria arborescens mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.55
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B4Z142|A0A1B4Z142_9VIRU Polyprotein OS=Alternaria arborescens mitovirus 1 OX=1826822 GN=RdRp PE=4 SV=1
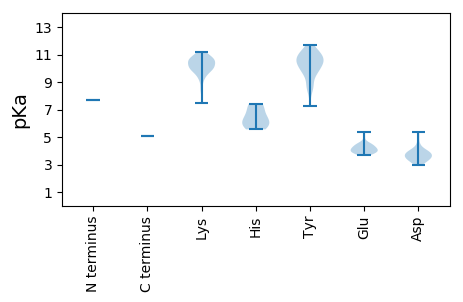
MM1 pKa = 7.71KK2 pKa = 10.21KK3 pKa = 9.72QLYY6 pKa = 9.72NITVKK11 pKa = 10.53LCSIVFPTINTLDD24 pKa = 3.56YY25 pKa = 10.76LNPYY29 pKa = 9.63FKK31 pKa = 11.02LLNRR35 pKa = 11.84LLNTQGLLKK44 pKa = 9.34TVKK47 pKa = 10.17YY48 pKa = 10.47LKK50 pKa = 9.76QCRR53 pKa = 11.84LHH55 pKa = 5.93CTRR58 pKa = 11.84YY59 pKa = 8.52MCGSPLLFNKK69 pKa = 10.25LKK71 pKa = 10.6IGLDD75 pKa = 3.14TDD77 pKa = 3.99GWPKK81 pKa = 10.66RR82 pKa = 11.84LDD84 pKa = 3.59FLKK87 pKa = 10.63PLAKK91 pKa = 10.42GSLEE95 pKa = 3.74QRR97 pKa = 11.84KK98 pKa = 8.45FLMTILCLSRR108 pKa = 11.84TLKK111 pKa = 11.16AEE113 pKa = 4.21GKK115 pKa = 9.14EE116 pKa = 3.88KK117 pKa = 11.07LKK119 pKa = 10.71IKK121 pKa = 10.07PDD123 pKa = 3.44YY124 pKa = 11.16DD125 pKa = 4.58SITKK129 pKa = 9.36PGRR132 pKa = 11.84IIKK135 pKa = 7.5TIPTGFIKK143 pKa = 10.6EE144 pKa = 4.11FVSNYY149 pKa = 7.24NLKK152 pKa = 9.09MEE154 pKa = 4.4KK155 pKa = 10.07PKK157 pKa = 11.03FEE159 pKa = 4.59INNIYY164 pKa = 10.32LSNKK168 pKa = 9.31AGPNGKK174 pKa = 8.33ATKK177 pKa = 8.69TAYY180 pKa = 10.3SSLLSYY186 pKa = 10.73SYY188 pKa = 11.69DD189 pKa = 3.26LMASLFKK196 pKa = 10.27ITDD199 pKa = 3.28QSGIDD204 pKa = 3.85YY205 pKa = 9.08FQSQYY210 pKa = 11.21NYY212 pKa = 9.94AWEE215 pKa = 4.42KK216 pKa = 10.86NFPSQKK222 pKa = 9.94LGKK225 pKa = 10.2LSFIYY230 pKa = 10.57DD231 pKa = 3.99PEE233 pKa = 5.39CKK235 pKa = 9.98LRR237 pKa = 11.84IVAIVDD243 pKa = 3.55YY244 pKa = 8.81YY245 pKa = 10.63TQLFLKK251 pKa = 9.58PIHH254 pKa = 6.02EE255 pKa = 5.03KK256 pKa = 10.46IMKK259 pKa = 9.78KK260 pKa = 10.0LQNLPCDD267 pKa = 3.41RR268 pKa = 11.84TYY270 pKa = 10.44TQSPLNEE277 pKa = 4.12WKK279 pKa = 10.5DD280 pKa = 3.74DD281 pKa = 3.79GNMFWSIDD289 pKa = 3.64LSSATDD295 pKa = 3.73RR296 pKa = 11.84FPISLQRR303 pKa = 11.84RR304 pKa = 11.84LLEE307 pKa = 3.68IAISKK312 pKa = 9.5EE313 pKa = 4.19VADD316 pKa = 4.0GWSFILSDD324 pKa = 4.22RR325 pKa = 11.84KK326 pKa = 10.6FEE328 pKa = 4.23TPEE331 pKa = 4.08GNLVQYY337 pKa = 8.75RR338 pKa = 11.84TGQPMGSYY346 pKa = 10.31SSWAAFTLTHH356 pKa = 6.43HH357 pKa = 7.27LVLHH361 pKa = 6.33WCAKK365 pKa = 10.56LNGIDD370 pKa = 3.91NFSDD374 pKa = 4.14YY375 pKa = 10.75IILGDD380 pKa = 5.33DD381 pKa = 3.38IVIKK385 pKa = 10.17NDD387 pKa = 2.96KK388 pKa = 9.71VARR391 pKa = 11.84TYY393 pKa = 10.58IKK395 pKa = 9.98WMDD398 pKa = 3.62YY399 pKa = 11.11LGVEE403 pKa = 4.3LSDD406 pKa = 3.97SKK408 pKa = 9.83THH410 pKa = 5.56VSKK413 pKa = 10.14DD414 pKa = 3.03TYY416 pKa = 10.94EE417 pKa = 3.89FAKK420 pKa = 10.36RR421 pKa = 11.84WFCKK425 pKa = 10.08GKK427 pKa = 10.14EE428 pKa = 4.15FTGLPMNGIVEE439 pKa = 4.51NIEE442 pKa = 3.8NPFIVMVNLYY452 pKa = 10.66DD453 pKa = 4.32FYY455 pKa = 11.25KK456 pKa = 11.13VKK458 pKa = 10.75GNYY461 pKa = 9.56LGSTKK466 pKa = 10.38NLPCILSSLYY476 pKa = 10.47KK477 pKa = 10.61GLSLKK482 pKa = 10.54LSKK485 pKa = 10.57KK486 pKa = 9.94FNNSRR491 pKa = 11.84FKK493 pKa = 9.77MKK495 pKa = 9.91IYY497 pKa = 9.79TFHH500 pKa = 7.43KK501 pKa = 10.38SLDD504 pKa = 3.51YY505 pKa = 11.18SFGYY509 pKa = 8.28LTYY512 pKa = 10.88DD513 pKa = 3.21SLRR516 pKa = 11.84EE517 pKa = 3.85LLCLNIKK524 pKa = 10.17NEE526 pKa = 3.86QFMIPDD532 pKa = 3.41EE533 pKa = 4.02QLIHH537 pKa = 5.72NTYY540 pKa = 11.11DD541 pKa = 3.43DD542 pKa = 3.83VVAQGMGGSVKK553 pKa = 10.84NSMTSLNNLASKK565 pKa = 10.59VIEE568 pKa = 4.23NKK570 pKa = 9.56TIYY573 pKa = 10.51NLEE576 pKa = 4.45DD577 pKa = 3.88PNEE580 pKa = 3.89LRR582 pKa = 11.84NYY584 pKa = 9.71PIFKK588 pKa = 10.67GIVNYY593 pKa = 10.1INNYY597 pKa = 8.62KK598 pKa = 10.75DD599 pKa = 4.98SVSKK603 pKa = 10.57WDD605 pKa = 3.77VNHH608 pKa = 6.85LNYY611 pKa = 9.88RR612 pKa = 11.84QKK614 pKa = 10.97SKK616 pKa = 10.91EE617 pKa = 3.96LLMLNIDD624 pKa = 3.56NVFGKK629 pKa = 9.9EE630 pKa = 3.82RR631 pKa = 11.84NKK633 pKa = 9.97TLEE636 pKa = 4.08LLNTGKK642 pKa = 10.05IFSLGFKK649 pKa = 10.14KK650 pKa = 10.48INEE653 pKa = 4.37TDD655 pKa = 3.31EE656 pKa = 3.95IMYY659 pKa = 10.18GSSIGEE665 pKa = 3.9STYY668 pKa = 11.2SYY670 pKa = 11.65NLDD673 pKa = 3.93LFNLIQNNYY682 pKa = 10.45SIDD685 pKa = 3.77LKK687 pKa = 10.94KK688 pKa = 10.83LKK690 pKa = 10.4EE691 pKa = 4.12LDD693 pKa = 3.07EE694 pKa = 4.52GTYY697 pKa = 10.05KK698 pKa = 10.81EE699 pKa = 4.5PVKK702 pKa = 9.75QTPASAYY709 pKa = 9.62DD710 pKa = 3.55AYY712 pKa = 11.04ANFFNN717 pKa = 5.07
MM1 pKa = 7.71KK2 pKa = 10.21KK3 pKa = 9.72QLYY6 pKa = 9.72NITVKK11 pKa = 10.53LCSIVFPTINTLDD24 pKa = 3.56YY25 pKa = 10.76LNPYY29 pKa = 9.63FKK31 pKa = 11.02LLNRR35 pKa = 11.84LLNTQGLLKK44 pKa = 9.34TVKK47 pKa = 10.17YY48 pKa = 10.47LKK50 pKa = 9.76QCRR53 pKa = 11.84LHH55 pKa = 5.93CTRR58 pKa = 11.84YY59 pKa = 8.52MCGSPLLFNKK69 pKa = 10.25LKK71 pKa = 10.6IGLDD75 pKa = 3.14TDD77 pKa = 3.99GWPKK81 pKa = 10.66RR82 pKa = 11.84LDD84 pKa = 3.59FLKK87 pKa = 10.63PLAKK91 pKa = 10.42GSLEE95 pKa = 3.74QRR97 pKa = 11.84KK98 pKa = 8.45FLMTILCLSRR108 pKa = 11.84TLKK111 pKa = 11.16AEE113 pKa = 4.21GKK115 pKa = 9.14EE116 pKa = 3.88KK117 pKa = 11.07LKK119 pKa = 10.71IKK121 pKa = 10.07PDD123 pKa = 3.44YY124 pKa = 11.16DD125 pKa = 4.58SITKK129 pKa = 9.36PGRR132 pKa = 11.84IIKK135 pKa = 7.5TIPTGFIKK143 pKa = 10.6EE144 pKa = 4.11FVSNYY149 pKa = 7.24NLKK152 pKa = 9.09MEE154 pKa = 4.4KK155 pKa = 10.07PKK157 pKa = 11.03FEE159 pKa = 4.59INNIYY164 pKa = 10.32LSNKK168 pKa = 9.31AGPNGKK174 pKa = 8.33ATKK177 pKa = 8.69TAYY180 pKa = 10.3SSLLSYY186 pKa = 10.73SYY188 pKa = 11.69DD189 pKa = 3.26LMASLFKK196 pKa = 10.27ITDD199 pKa = 3.28QSGIDD204 pKa = 3.85YY205 pKa = 9.08FQSQYY210 pKa = 11.21NYY212 pKa = 9.94AWEE215 pKa = 4.42KK216 pKa = 10.86NFPSQKK222 pKa = 9.94LGKK225 pKa = 10.2LSFIYY230 pKa = 10.57DD231 pKa = 3.99PEE233 pKa = 5.39CKK235 pKa = 9.98LRR237 pKa = 11.84IVAIVDD243 pKa = 3.55YY244 pKa = 8.81YY245 pKa = 10.63TQLFLKK251 pKa = 9.58PIHH254 pKa = 6.02EE255 pKa = 5.03KK256 pKa = 10.46IMKK259 pKa = 9.78KK260 pKa = 10.0LQNLPCDD267 pKa = 3.41RR268 pKa = 11.84TYY270 pKa = 10.44TQSPLNEE277 pKa = 4.12WKK279 pKa = 10.5DD280 pKa = 3.74DD281 pKa = 3.79GNMFWSIDD289 pKa = 3.64LSSATDD295 pKa = 3.73RR296 pKa = 11.84FPISLQRR303 pKa = 11.84RR304 pKa = 11.84LLEE307 pKa = 3.68IAISKK312 pKa = 9.5EE313 pKa = 4.19VADD316 pKa = 4.0GWSFILSDD324 pKa = 4.22RR325 pKa = 11.84KK326 pKa = 10.6FEE328 pKa = 4.23TPEE331 pKa = 4.08GNLVQYY337 pKa = 8.75RR338 pKa = 11.84TGQPMGSYY346 pKa = 10.31SSWAAFTLTHH356 pKa = 6.43HH357 pKa = 7.27LVLHH361 pKa = 6.33WCAKK365 pKa = 10.56LNGIDD370 pKa = 3.91NFSDD374 pKa = 4.14YY375 pKa = 10.75IILGDD380 pKa = 5.33DD381 pKa = 3.38IVIKK385 pKa = 10.17NDD387 pKa = 2.96KK388 pKa = 9.71VARR391 pKa = 11.84TYY393 pKa = 10.58IKK395 pKa = 9.98WMDD398 pKa = 3.62YY399 pKa = 11.11LGVEE403 pKa = 4.3LSDD406 pKa = 3.97SKK408 pKa = 9.83THH410 pKa = 5.56VSKK413 pKa = 10.14DD414 pKa = 3.03TYY416 pKa = 10.94EE417 pKa = 3.89FAKK420 pKa = 10.36RR421 pKa = 11.84WFCKK425 pKa = 10.08GKK427 pKa = 10.14EE428 pKa = 4.15FTGLPMNGIVEE439 pKa = 4.51NIEE442 pKa = 3.8NPFIVMVNLYY452 pKa = 10.66DD453 pKa = 4.32FYY455 pKa = 11.25KK456 pKa = 11.13VKK458 pKa = 10.75GNYY461 pKa = 9.56LGSTKK466 pKa = 10.38NLPCILSSLYY476 pKa = 10.47KK477 pKa = 10.61GLSLKK482 pKa = 10.54LSKK485 pKa = 10.57KK486 pKa = 9.94FNNSRR491 pKa = 11.84FKK493 pKa = 9.77MKK495 pKa = 9.91IYY497 pKa = 9.79TFHH500 pKa = 7.43KK501 pKa = 10.38SLDD504 pKa = 3.51YY505 pKa = 11.18SFGYY509 pKa = 8.28LTYY512 pKa = 10.88DD513 pKa = 3.21SLRR516 pKa = 11.84EE517 pKa = 3.85LLCLNIKK524 pKa = 10.17NEE526 pKa = 3.86QFMIPDD532 pKa = 3.41EE533 pKa = 4.02QLIHH537 pKa = 5.72NTYY540 pKa = 11.11DD541 pKa = 3.43DD542 pKa = 3.83VVAQGMGGSVKK553 pKa = 10.84NSMTSLNNLASKK565 pKa = 10.59VIEE568 pKa = 4.23NKK570 pKa = 9.56TIYY573 pKa = 10.51NLEE576 pKa = 4.45DD577 pKa = 3.88PNEE580 pKa = 3.89LRR582 pKa = 11.84NYY584 pKa = 9.71PIFKK588 pKa = 10.67GIVNYY593 pKa = 10.1INNYY597 pKa = 8.62KK598 pKa = 10.75DD599 pKa = 4.98SVSKK603 pKa = 10.57WDD605 pKa = 3.77VNHH608 pKa = 6.85LNYY611 pKa = 9.88RR612 pKa = 11.84QKK614 pKa = 10.97SKK616 pKa = 10.91EE617 pKa = 3.96LLMLNIDD624 pKa = 3.56NVFGKK629 pKa = 9.9EE630 pKa = 3.82RR631 pKa = 11.84NKK633 pKa = 9.97TLEE636 pKa = 4.08LLNTGKK642 pKa = 10.05IFSLGFKK649 pKa = 10.14KK650 pKa = 10.48INEE653 pKa = 4.37TDD655 pKa = 3.31EE656 pKa = 3.95IMYY659 pKa = 10.18GSSIGEE665 pKa = 3.9STYY668 pKa = 11.2SYY670 pKa = 11.65NLDD673 pKa = 3.93LFNLIQNNYY682 pKa = 10.45SIDD685 pKa = 3.77LKK687 pKa = 10.94KK688 pKa = 10.83LKK690 pKa = 10.4EE691 pKa = 4.12LDD693 pKa = 3.07EE694 pKa = 4.52GTYY697 pKa = 10.05KK698 pKa = 10.81EE699 pKa = 4.5PVKK702 pKa = 9.75QTPASAYY709 pKa = 9.62DD710 pKa = 3.55AYY712 pKa = 11.04ANFFNN717 pKa = 5.07
Molecular weight: 83.14 kDa
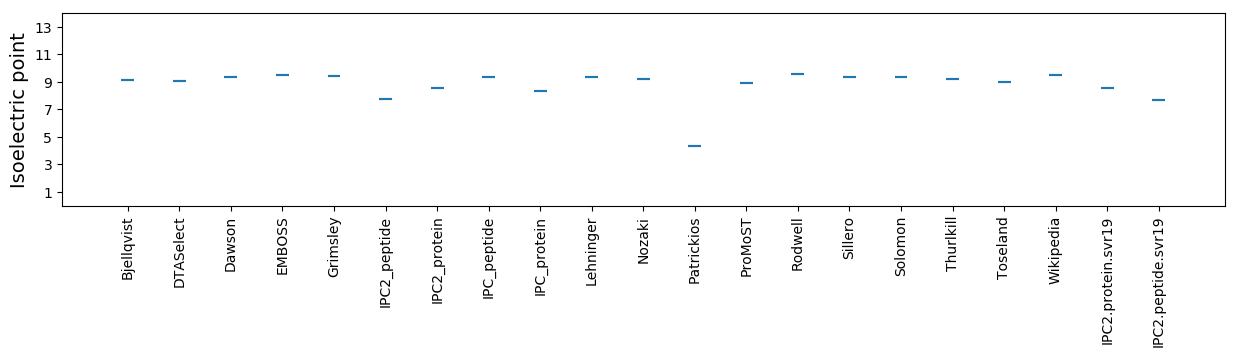
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B4Z142|A0A1B4Z142_9VIRU Polyprotein OS=Alternaria arborescens mitovirus 1 OX=1826822 GN=RdRp PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 10.21KK3 pKa = 9.72QLYY6 pKa = 9.72NITVKK11 pKa = 10.53LCSIVFPTINTLDD24 pKa = 3.56YY25 pKa = 10.76LNPYY29 pKa = 9.63FKK31 pKa = 11.02LLNRR35 pKa = 11.84LLNTQGLLKK44 pKa = 9.34TVKK47 pKa = 10.17YY48 pKa = 10.47LKK50 pKa = 9.76QCRR53 pKa = 11.84LHH55 pKa = 5.93CTRR58 pKa = 11.84YY59 pKa = 8.52MCGSPLLFNKK69 pKa = 10.25LKK71 pKa = 10.6IGLDD75 pKa = 3.14TDD77 pKa = 3.99GWPKK81 pKa = 10.66RR82 pKa = 11.84LDD84 pKa = 3.59FLKK87 pKa = 10.63PLAKK91 pKa = 10.42GSLEE95 pKa = 3.74QRR97 pKa = 11.84KK98 pKa = 8.45FLMTILCLSRR108 pKa = 11.84TLKK111 pKa = 11.16AEE113 pKa = 4.21GKK115 pKa = 9.14EE116 pKa = 3.88KK117 pKa = 11.07LKK119 pKa = 10.71IKK121 pKa = 10.07PDD123 pKa = 3.44YY124 pKa = 11.16DD125 pKa = 4.58SITKK129 pKa = 9.36PGRR132 pKa = 11.84IIKK135 pKa = 7.5TIPTGFIKK143 pKa = 10.6EE144 pKa = 4.11FVSNYY149 pKa = 7.24NLKK152 pKa = 9.09MEE154 pKa = 4.4KK155 pKa = 10.07PKK157 pKa = 11.03FEE159 pKa = 4.59INNIYY164 pKa = 10.32LSNKK168 pKa = 9.31AGPNGKK174 pKa = 8.33ATKK177 pKa = 8.69TAYY180 pKa = 10.3SSLLSYY186 pKa = 10.73SYY188 pKa = 11.69DD189 pKa = 3.26LMASLFKK196 pKa = 10.27ITDD199 pKa = 3.28QSGIDD204 pKa = 3.85YY205 pKa = 9.08FQSQYY210 pKa = 11.21NYY212 pKa = 9.94AWEE215 pKa = 4.42KK216 pKa = 10.86NFPSQKK222 pKa = 9.94LGKK225 pKa = 10.2LSFIYY230 pKa = 10.57DD231 pKa = 3.99PEE233 pKa = 5.39CKK235 pKa = 9.98LRR237 pKa = 11.84IVAIVDD243 pKa = 3.55YY244 pKa = 8.81YY245 pKa = 10.63TQLFLKK251 pKa = 9.58PIHH254 pKa = 6.02EE255 pKa = 5.03KK256 pKa = 10.46IMKK259 pKa = 9.78KK260 pKa = 10.0LQNLPCDD267 pKa = 3.41RR268 pKa = 11.84TYY270 pKa = 10.44TQSPLNEE277 pKa = 4.12WKK279 pKa = 10.5DD280 pKa = 3.74DD281 pKa = 3.79GNMFWSIDD289 pKa = 3.64LSSATDD295 pKa = 3.73RR296 pKa = 11.84FPISLQRR303 pKa = 11.84RR304 pKa = 11.84LLEE307 pKa = 3.68IAISKK312 pKa = 9.5EE313 pKa = 4.19VADD316 pKa = 4.0GWSFILSDD324 pKa = 4.22RR325 pKa = 11.84KK326 pKa = 10.6FEE328 pKa = 4.23TPEE331 pKa = 4.08GNLVQYY337 pKa = 8.75RR338 pKa = 11.84TGQPMGSYY346 pKa = 10.31SSWAAFTLTHH356 pKa = 6.43HH357 pKa = 7.27LVLHH361 pKa = 6.33WCAKK365 pKa = 10.56LNGIDD370 pKa = 3.91NFSDD374 pKa = 4.14YY375 pKa = 10.75IILGDD380 pKa = 5.33DD381 pKa = 3.38IVIKK385 pKa = 10.17NDD387 pKa = 2.96KK388 pKa = 9.71VARR391 pKa = 11.84TYY393 pKa = 10.58IKK395 pKa = 9.98WMDD398 pKa = 3.62YY399 pKa = 11.11LGVEE403 pKa = 4.3LSDD406 pKa = 3.97SKK408 pKa = 9.83THH410 pKa = 5.56VSKK413 pKa = 10.14DD414 pKa = 3.03TYY416 pKa = 10.94EE417 pKa = 3.89FAKK420 pKa = 10.36RR421 pKa = 11.84WFCKK425 pKa = 10.08GKK427 pKa = 10.14EE428 pKa = 4.15FTGLPMNGIVEE439 pKa = 4.51NIEE442 pKa = 3.8NPFIVMVNLYY452 pKa = 10.66DD453 pKa = 4.32FYY455 pKa = 11.25KK456 pKa = 11.13VKK458 pKa = 10.75GNYY461 pKa = 9.56LGSTKK466 pKa = 10.38NLPCILSSLYY476 pKa = 10.47KK477 pKa = 10.61GLSLKK482 pKa = 10.54LSKK485 pKa = 10.57KK486 pKa = 9.94FNNSRR491 pKa = 11.84FKK493 pKa = 9.77MKK495 pKa = 9.91IYY497 pKa = 9.79TFHH500 pKa = 7.43KK501 pKa = 10.38SLDD504 pKa = 3.51YY505 pKa = 11.18SFGYY509 pKa = 8.28LTYY512 pKa = 10.88DD513 pKa = 3.21SLRR516 pKa = 11.84EE517 pKa = 3.85LLCLNIKK524 pKa = 10.17NEE526 pKa = 3.86QFMIPDD532 pKa = 3.41EE533 pKa = 4.02QLIHH537 pKa = 5.72NTYY540 pKa = 11.11DD541 pKa = 3.43DD542 pKa = 3.83VVAQGMGGSVKK553 pKa = 10.84NSMTSLNNLASKK565 pKa = 10.59VIEE568 pKa = 4.23NKK570 pKa = 9.56TIYY573 pKa = 10.51NLEE576 pKa = 4.45DD577 pKa = 3.88PNEE580 pKa = 3.89LRR582 pKa = 11.84NYY584 pKa = 9.71PIFKK588 pKa = 10.67GIVNYY593 pKa = 10.1INNYY597 pKa = 8.62KK598 pKa = 10.75DD599 pKa = 4.98SVSKK603 pKa = 10.57WDD605 pKa = 3.77VNHH608 pKa = 6.85LNYY611 pKa = 9.88RR612 pKa = 11.84QKK614 pKa = 10.97SKK616 pKa = 10.91EE617 pKa = 3.96LLMLNIDD624 pKa = 3.56NVFGKK629 pKa = 9.9EE630 pKa = 3.82RR631 pKa = 11.84NKK633 pKa = 9.97TLEE636 pKa = 4.08LLNTGKK642 pKa = 10.05IFSLGFKK649 pKa = 10.14KK650 pKa = 10.48INEE653 pKa = 4.37TDD655 pKa = 3.31EE656 pKa = 3.95IMYY659 pKa = 10.18GSSIGEE665 pKa = 3.9STYY668 pKa = 11.2SYY670 pKa = 11.65NLDD673 pKa = 3.93LFNLIQNNYY682 pKa = 10.45SIDD685 pKa = 3.77LKK687 pKa = 10.94KK688 pKa = 10.83LKK690 pKa = 10.4EE691 pKa = 4.12LDD693 pKa = 3.07EE694 pKa = 4.52GTYY697 pKa = 10.05KK698 pKa = 10.81EE699 pKa = 4.5PVKK702 pKa = 9.75QTPASAYY709 pKa = 9.62DD710 pKa = 3.55AYY712 pKa = 11.04ANFFNN717 pKa = 5.07
MM1 pKa = 7.71KK2 pKa = 10.21KK3 pKa = 9.72QLYY6 pKa = 9.72NITVKK11 pKa = 10.53LCSIVFPTINTLDD24 pKa = 3.56YY25 pKa = 10.76LNPYY29 pKa = 9.63FKK31 pKa = 11.02LLNRR35 pKa = 11.84LLNTQGLLKK44 pKa = 9.34TVKK47 pKa = 10.17YY48 pKa = 10.47LKK50 pKa = 9.76QCRR53 pKa = 11.84LHH55 pKa = 5.93CTRR58 pKa = 11.84YY59 pKa = 8.52MCGSPLLFNKK69 pKa = 10.25LKK71 pKa = 10.6IGLDD75 pKa = 3.14TDD77 pKa = 3.99GWPKK81 pKa = 10.66RR82 pKa = 11.84LDD84 pKa = 3.59FLKK87 pKa = 10.63PLAKK91 pKa = 10.42GSLEE95 pKa = 3.74QRR97 pKa = 11.84KK98 pKa = 8.45FLMTILCLSRR108 pKa = 11.84TLKK111 pKa = 11.16AEE113 pKa = 4.21GKK115 pKa = 9.14EE116 pKa = 3.88KK117 pKa = 11.07LKK119 pKa = 10.71IKK121 pKa = 10.07PDD123 pKa = 3.44YY124 pKa = 11.16DD125 pKa = 4.58SITKK129 pKa = 9.36PGRR132 pKa = 11.84IIKK135 pKa = 7.5TIPTGFIKK143 pKa = 10.6EE144 pKa = 4.11FVSNYY149 pKa = 7.24NLKK152 pKa = 9.09MEE154 pKa = 4.4KK155 pKa = 10.07PKK157 pKa = 11.03FEE159 pKa = 4.59INNIYY164 pKa = 10.32LSNKK168 pKa = 9.31AGPNGKK174 pKa = 8.33ATKK177 pKa = 8.69TAYY180 pKa = 10.3SSLLSYY186 pKa = 10.73SYY188 pKa = 11.69DD189 pKa = 3.26LMASLFKK196 pKa = 10.27ITDD199 pKa = 3.28QSGIDD204 pKa = 3.85YY205 pKa = 9.08FQSQYY210 pKa = 11.21NYY212 pKa = 9.94AWEE215 pKa = 4.42KK216 pKa = 10.86NFPSQKK222 pKa = 9.94LGKK225 pKa = 10.2LSFIYY230 pKa = 10.57DD231 pKa = 3.99PEE233 pKa = 5.39CKK235 pKa = 9.98LRR237 pKa = 11.84IVAIVDD243 pKa = 3.55YY244 pKa = 8.81YY245 pKa = 10.63TQLFLKK251 pKa = 9.58PIHH254 pKa = 6.02EE255 pKa = 5.03KK256 pKa = 10.46IMKK259 pKa = 9.78KK260 pKa = 10.0LQNLPCDD267 pKa = 3.41RR268 pKa = 11.84TYY270 pKa = 10.44TQSPLNEE277 pKa = 4.12WKK279 pKa = 10.5DD280 pKa = 3.74DD281 pKa = 3.79GNMFWSIDD289 pKa = 3.64LSSATDD295 pKa = 3.73RR296 pKa = 11.84FPISLQRR303 pKa = 11.84RR304 pKa = 11.84LLEE307 pKa = 3.68IAISKK312 pKa = 9.5EE313 pKa = 4.19VADD316 pKa = 4.0GWSFILSDD324 pKa = 4.22RR325 pKa = 11.84KK326 pKa = 10.6FEE328 pKa = 4.23TPEE331 pKa = 4.08GNLVQYY337 pKa = 8.75RR338 pKa = 11.84TGQPMGSYY346 pKa = 10.31SSWAAFTLTHH356 pKa = 6.43HH357 pKa = 7.27LVLHH361 pKa = 6.33WCAKK365 pKa = 10.56LNGIDD370 pKa = 3.91NFSDD374 pKa = 4.14YY375 pKa = 10.75IILGDD380 pKa = 5.33DD381 pKa = 3.38IVIKK385 pKa = 10.17NDD387 pKa = 2.96KK388 pKa = 9.71VARR391 pKa = 11.84TYY393 pKa = 10.58IKK395 pKa = 9.98WMDD398 pKa = 3.62YY399 pKa = 11.11LGVEE403 pKa = 4.3LSDD406 pKa = 3.97SKK408 pKa = 9.83THH410 pKa = 5.56VSKK413 pKa = 10.14DD414 pKa = 3.03TYY416 pKa = 10.94EE417 pKa = 3.89FAKK420 pKa = 10.36RR421 pKa = 11.84WFCKK425 pKa = 10.08GKK427 pKa = 10.14EE428 pKa = 4.15FTGLPMNGIVEE439 pKa = 4.51NIEE442 pKa = 3.8NPFIVMVNLYY452 pKa = 10.66DD453 pKa = 4.32FYY455 pKa = 11.25KK456 pKa = 11.13VKK458 pKa = 10.75GNYY461 pKa = 9.56LGSTKK466 pKa = 10.38NLPCILSSLYY476 pKa = 10.47KK477 pKa = 10.61GLSLKK482 pKa = 10.54LSKK485 pKa = 10.57KK486 pKa = 9.94FNNSRR491 pKa = 11.84FKK493 pKa = 9.77MKK495 pKa = 9.91IYY497 pKa = 9.79TFHH500 pKa = 7.43KK501 pKa = 10.38SLDD504 pKa = 3.51YY505 pKa = 11.18SFGYY509 pKa = 8.28LTYY512 pKa = 10.88DD513 pKa = 3.21SLRR516 pKa = 11.84EE517 pKa = 3.85LLCLNIKK524 pKa = 10.17NEE526 pKa = 3.86QFMIPDD532 pKa = 3.41EE533 pKa = 4.02QLIHH537 pKa = 5.72NTYY540 pKa = 11.11DD541 pKa = 3.43DD542 pKa = 3.83VVAQGMGGSVKK553 pKa = 10.84NSMTSLNNLASKK565 pKa = 10.59VIEE568 pKa = 4.23NKK570 pKa = 9.56TIYY573 pKa = 10.51NLEE576 pKa = 4.45DD577 pKa = 3.88PNEE580 pKa = 3.89LRR582 pKa = 11.84NYY584 pKa = 9.71PIFKK588 pKa = 10.67GIVNYY593 pKa = 10.1INNYY597 pKa = 8.62KK598 pKa = 10.75DD599 pKa = 4.98SVSKK603 pKa = 10.57WDD605 pKa = 3.77VNHH608 pKa = 6.85LNYY611 pKa = 9.88RR612 pKa = 11.84QKK614 pKa = 10.97SKK616 pKa = 10.91EE617 pKa = 3.96LLMLNIDD624 pKa = 3.56NVFGKK629 pKa = 9.9EE630 pKa = 3.82RR631 pKa = 11.84NKK633 pKa = 9.97TLEE636 pKa = 4.08LLNTGKK642 pKa = 10.05IFSLGFKK649 pKa = 10.14KK650 pKa = 10.48INEE653 pKa = 4.37TDD655 pKa = 3.31EE656 pKa = 3.95IMYY659 pKa = 10.18GSSIGEE665 pKa = 3.9STYY668 pKa = 11.2SYY670 pKa = 11.65NLDD673 pKa = 3.93LFNLIQNNYY682 pKa = 10.45SIDD685 pKa = 3.77LKK687 pKa = 10.94KK688 pKa = 10.83LKK690 pKa = 10.4EE691 pKa = 4.12LDD693 pKa = 3.07EE694 pKa = 4.52GTYY697 pKa = 10.05KK698 pKa = 10.81EE699 pKa = 4.5PVKK702 pKa = 9.75QTPASAYY709 pKa = 9.62DD710 pKa = 3.55AYY712 pKa = 11.04ANFFNN717 pKa = 5.07
Molecular weight: 83.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
717 |
717 |
717 |
717.0 |
83.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.068 ± 0.0 | 1.534 ± 0.0 |
5.718 ± 0.0 | 4.742 ± 0.0 |
5.021 ± 0.0 | 5.16 ± 0.0 |
1.255 ± 0.0 | 7.113 ± 0.0 |
10.739 ± 0.0 | 11.994 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.371 ± 0.0 | 7.671 ± 0.0 |
3.626 ± 0.0 | 2.789 ± 0.0 |
2.929 ± 0.0 | 7.531 ± 0.0 |
5.439 ± 0.0 | 3.626 ± 0.0 |
1.395 ± 0.0 | 6.276 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
