
Avon-Heathcote Estuary associated circular virus 10
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins
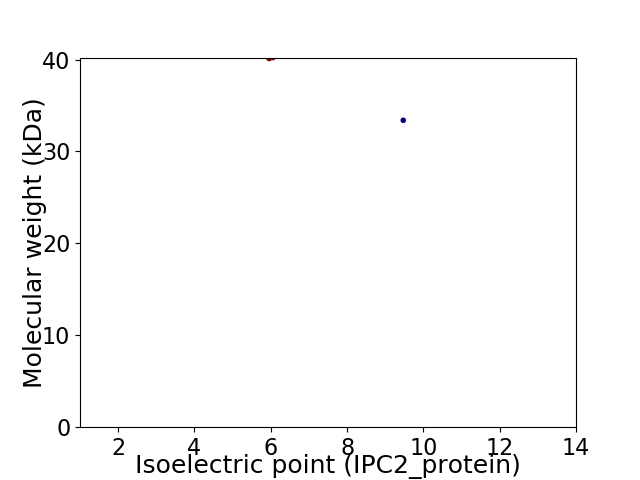
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IBL9|A0A0C5IBL9_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 10 OX=1618233 PE=3 SV=1
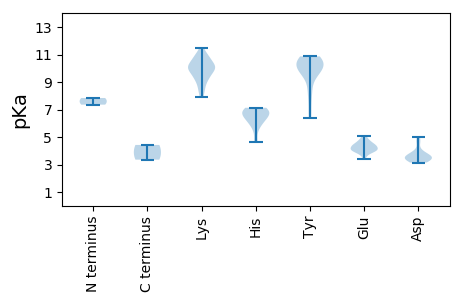
MM1 pKa = 7.84PVTQPVTKK9 pKa = 9.21ATRR12 pKa = 11.84AICFTEE18 pKa = 4.97FKK20 pKa = 10.93KK21 pKa = 9.75LTNWDD26 pKa = 3.68DD27 pKa = 3.5VKK29 pKa = 11.44SQLRR33 pKa = 11.84YY34 pKa = 8.63YY35 pKa = 10.81AYY37 pKa = 10.37AEE39 pKa = 4.19EE40 pKa = 4.53TCPKK44 pKa = 9.18TGKK47 pKa = 9.49KK48 pKa = 9.54HH49 pKa = 4.62YY50 pKa = 10.29QGFAYY55 pKa = 9.52SWKK58 pKa = 9.91PIRR61 pKa = 11.84FSGWKK66 pKa = 10.13KK67 pKa = 9.21MFPTAHH73 pKa = 6.45IEE75 pKa = 3.86QMRR78 pKa = 11.84GNFRR82 pKa = 11.84EE83 pKa = 3.94NTAYY87 pKa = 10.38CSKK90 pKa = 10.12EE91 pKa = 3.41GSLIEE96 pKa = 4.74FGEE99 pKa = 4.3KK100 pKa = 9.98PNEE103 pKa = 4.0NGVQGTAIEE112 pKa = 4.1YY113 pKa = 9.97KK114 pKa = 10.28RR115 pKa = 11.84RR116 pKa = 11.84IEE118 pKa = 4.21EE119 pKa = 4.25GQSTMEE125 pKa = 3.87IAEE128 pKa = 4.43DD129 pKa = 3.72EE130 pKa = 4.44NLFGQYY136 pKa = 9.63LHH138 pKa = 7.13SYY140 pKa = 9.33SALEE144 pKa = 4.45KK145 pKa = 10.44YY146 pKa = 8.01EE147 pKa = 3.59QHH149 pKa = 6.7IRR151 pKa = 11.84GKK153 pKa = 10.2KK154 pKa = 8.87IKK156 pKa = 10.34LDD158 pKa = 3.52EE159 pKa = 4.65TPPEE163 pKa = 4.15VYY165 pKa = 10.07IRR167 pKa = 11.84WGAAGTGKK175 pKa = 7.89THH177 pKa = 6.96GVFKK181 pKa = 10.72RR182 pKa = 11.84YY183 pKa = 9.84GRR185 pKa = 11.84DD186 pKa = 3.14KK187 pKa = 11.36VCIVPDD193 pKa = 3.67NTGKK197 pKa = 9.76WFDD200 pKa = 3.51NCHH203 pKa = 6.2NSDD206 pKa = 3.24VMLFDD211 pKa = 5.0DD212 pKa = 4.75VEE214 pKa = 4.77IDD216 pKa = 3.53AVPPISQFLRR226 pKa = 11.84LTDD229 pKa = 4.52RR230 pKa = 11.84YY231 pKa = 10.06PMQVPVKK238 pKa = 10.65GGFIWWKK245 pKa = 8.36PKK247 pKa = 10.8VIVFTSNLPWEE258 pKa = 4.51QWWNTLNTEE267 pKa = 4.24HH268 pKa = 6.84KK269 pKa = 10.13RR270 pKa = 11.84AVQRR274 pKa = 11.84RR275 pKa = 11.84LTRR278 pKa = 11.84VTRR281 pKa = 11.84IYY283 pKa = 10.59KK284 pKa = 9.71DD285 pKa = 3.39HH286 pKa = 7.06EE287 pKa = 4.02KK288 pKa = 10.96VEE290 pKa = 4.34YY291 pKa = 10.74EE292 pKa = 3.89NANYY296 pKa = 10.0EE297 pKa = 4.71LLSQTQGFFASPPQLQQEE315 pKa = 4.53VQEE318 pKa = 4.71EE319 pKa = 4.4PVHH322 pKa = 5.7SQEE325 pKa = 4.5EE326 pKa = 5.08GEE328 pKa = 4.39QVHH331 pKa = 6.12EE332 pKa = 4.37EE333 pKa = 4.03SGSASEE339 pKa = 4.11EE340 pKa = 4.03HH341 pKa = 6.71SAVV344 pKa = 3.35
MM1 pKa = 7.84PVTQPVTKK9 pKa = 9.21ATRR12 pKa = 11.84AICFTEE18 pKa = 4.97FKK20 pKa = 10.93KK21 pKa = 9.75LTNWDD26 pKa = 3.68DD27 pKa = 3.5VKK29 pKa = 11.44SQLRR33 pKa = 11.84YY34 pKa = 8.63YY35 pKa = 10.81AYY37 pKa = 10.37AEE39 pKa = 4.19EE40 pKa = 4.53TCPKK44 pKa = 9.18TGKK47 pKa = 9.49KK48 pKa = 9.54HH49 pKa = 4.62YY50 pKa = 10.29QGFAYY55 pKa = 9.52SWKK58 pKa = 9.91PIRR61 pKa = 11.84FSGWKK66 pKa = 10.13KK67 pKa = 9.21MFPTAHH73 pKa = 6.45IEE75 pKa = 3.86QMRR78 pKa = 11.84GNFRR82 pKa = 11.84EE83 pKa = 3.94NTAYY87 pKa = 10.38CSKK90 pKa = 10.12EE91 pKa = 3.41GSLIEE96 pKa = 4.74FGEE99 pKa = 4.3KK100 pKa = 9.98PNEE103 pKa = 4.0NGVQGTAIEE112 pKa = 4.1YY113 pKa = 9.97KK114 pKa = 10.28RR115 pKa = 11.84RR116 pKa = 11.84IEE118 pKa = 4.21EE119 pKa = 4.25GQSTMEE125 pKa = 3.87IAEE128 pKa = 4.43DD129 pKa = 3.72EE130 pKa = 4.44NLFGQYY136 pKa = 9.63LHH138 pKa = 7.13SYY140 pKa = 9.33SALEE144 pKa = 4.45KK145 pKa = 10.44YY146 pKa = 8.01EE147 pKa = 3.59QHH149 pKa = 6.7IRR151 pKa = 11.84GKK153 pKa = 10.2KK154 pKa = 8.87IKK156 pKa = 10.34LDD158 pKa = 3.52EE159 pKa = 4.65TPPEE163 pKa = 4.15VYY165 pKa = 10.07IRR167 pKa = 11.84WGAAGTGKK175 pKa = 7.89THH177 pKa = 6.96GVFKK181 pKa = 10.72RR182 pKa = 11.84YY183 pKa = 9.84GRR185 pKa = 11.84DD186 pKa = 3.14KK187 pKa = 11.36VCIVPDD193 pKa = 3.67NTGKK197 pKa = 9.76WFDD200 pKa = 3.51NCHH203 pKa = 6.2NSDD206 pKa = 3.24VMLFDD211 pKa = 5.0DD212 pKa = 4.75VEE214 pKa = 4.77IDD216 pKa = 3.53AVPPISQFLRR226 pKa = 11.84LTDD229 pKa = 4.52RR230 pKa = 11.84YY231 pKa = 10.06PMQVPVKK238 pKa = 10.65GGFIWWKK245 pKa = 8.36PKK247 pKa = 10.8VIVFTSNLPWEE258 pKa = 4.51QWWNTLNTEE267 pKa = 4.24HH268 pKa = 6.84KK269 pKa = 10.13RR270 pKa = 11.84AVQRR274 pKa = 11.84RR275 pKa = 11.84LTRR278 pKa = 11.84VTRR281 pKa = 11.84IYY283 pKa = 10.59KK284 pKa = 9.71DD285 pKa = 3.39HH286 pKa = 7.06EE287 pKa = 4.02KK288 pKa = 10.96VEE290 pKa = 4.34YY291 pKa = 10.74EE292 pKa = 3.89NANYY296 pKa = 10.0EE297 pKa = 4.71LLSQTQGFFASPPQLQQEE315 pKa = 4.53VQEE318 pKa = 4.71EE319 pKa = 4.4PVHH322 pKa = 5.7SQEE325 pKa = 4.5EE326 pKa = 5.08GEE328 pKa = 4.39QVHH331 pKa = 6.12EE332 pKa = 4.37EE333 pKa = 4.03SGSASEE339 pKa = 4.11EE340 pKa = 4.03HH341 pKa = 6.71SAVV344 pKa = 3.35
Molecular weight: 40.11 kDa
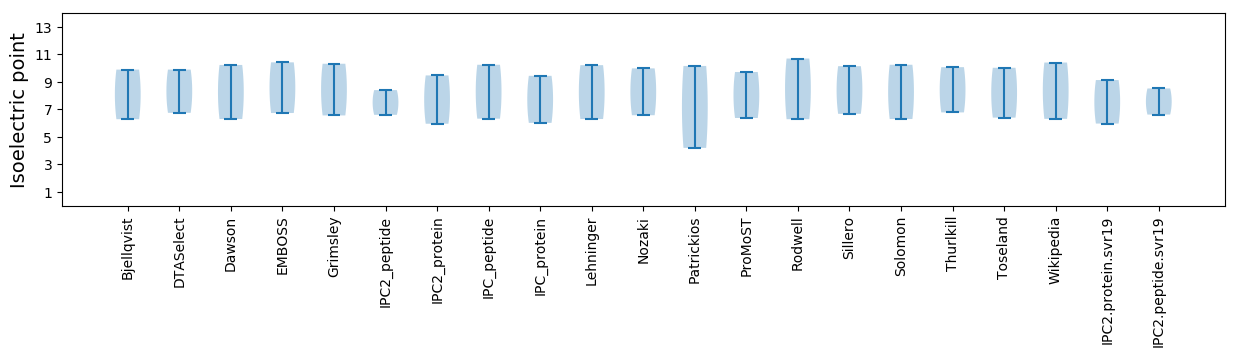
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IBL9|A0A0C5IBL9_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 10 OX=1618233 PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 10.09KK3 pKa = 9.55WNTKK7 pKa = 8.58MPITSYY13 pKa = 9.85FRR15 pKa = 11.84KK16 pKa = 9.88RR17 pKa = 11.84RR18 pKa = 11.84ASSQARR24 pKa = 11.84PNYY27 pKa = 9.77SKK29 pKa = 10.9KK30 pKa = 10.41SKK32 pKa = 10.13KK33 pKa = 10.0NPYY36 pKa = 6.4TRR38 pKa = 11.84KK39 pKa = 9.87RR40 pKa = 11.84KK41 pKa = 8.94ASKK44 pKa = 9.18YY45 pKa = 6.8MKK47 pKa = 10.13RR48 pKa = 11.84AVAQAKK54 pKa = 8.4STARR58 pKa = 11.84FNTQVKK64 pKa = 10.05NVMLAMEE71 pKa = 4.16EE72 pKa = 4.32TKK74 pKa = 11.08YY75 pKa = 9.66KK76 pKa = 7.95TTNLTGLPAYY86 pKa = 7.6ATTMYY91 pKa = 10.35HH92 pKa = 6.78NKK94 pKa = 8.49LTSFKK99 pKa = 10.56LWEE102 pKa = 4.12QGSNGTMALFPLQRR116 pKa = 11.84GTDD119 pKa = 3.16QDD121 pKa = 4.02EE122 pKa = 4.35IVGRR126 pKa = 11.84EE127 pKa = 3.99YY128 pKa = 10.86YY129 pKa = 10.77AKK131 pKa = 10.25SLKK134 pKa = 9.72IDD136 pKa = 3.55LQFTFPYY143 pKa = 10.26DD144 pKa = 3.51RR145 pKa = 11.84LGSTIKK151 pKa = 10.14VWYY154 pKa = 9.9VPVKK158 pKa = 9.89TGQPEE163 pKa = 3.64PAYY166 pKa = 10.71DD167 pKa = 3.37EE168 pKa = 4.77FFRR171 pKa = 11.84NTSGNIMLDD180 pKa = 3.27PRR182 pKa = 11.84NMQNYY187 pKa = 8.58PYY189 pKa = 10.6AKK191 pKa = 9.13YY192 pKa = 10.28LGSFRR197 pKa = 11.84PRR199 pKa = 11.84VKK201 pKa = 9.45QTTFGTLVNGNGVLGGRR218 pKa = 11.84DD219 pKa = 3.23ATVVKK224 pKa = 10.17SYY226 pKa = 10.61RR227 pKa = 11.84ISFNKK232 pKa = 9.41FFKK235 pKa = 10.62LRR237 pKa = 11.84SNVGGDD243 pKa = 3.64AEE245 pKa = 5.06IDD247 pKa = 3.86FNDD250 pKa = 3.94IPNLAEE256 pKa = 3.84KK257 pKa = 10.77AFLVFTAYY265 pKa = 10.63NEE267 pKa = 4.03EE268 pKa = 4.15AALEE272 pKa = 4.04TDD274 pKa = 3.54QVISNVEE281 pKa = 3.84GAFTFSYY288 pKa = 10.84KK289 pKa = 10.81DD290 pKa = 3.24PP291 pKa = 4.43
MM1 pKa = 7.36KK2 pKa = 10.09KK3 pKa = 9.55WNTKK7 pKa = 8.58MPITSYY13 pKa = 9.85FRR15 pKa = 11.84KK16 pKa = 9.88RR17 pKa = 11.84RR18 pKa = 11.84ASSQARR24 pKa = 11.84PNYY27 pKa = 9.77SKK29 pKa = 10.9KK30 pKa = 10.41SKK32 pKa = 10.13KK33 pKa = 10.0NPYY36 pKa = 6.4TRR38 pKa = 11.84KK39 pKa = 9.87RR40 pKa = 11.84KK41 pKa = 8.94ASKK44 pKa = 9.18YY45 pKa = 6.8MKK47 pKa = 10.13RR48 pKa = 11.84AVAQAKK54 pKa = 8.4STARR58 pKa = 11.84FNTQVKK64 pKa = 10.05NVMLAMEE71 pKa = 4.16EE72 pKa = 4.32TKK74 pKa = 11.08YY75 pKa = 9.66KK76 pKa = 7.95TTNLTGLPAYY86 pKa = 7.6ATTMYY91 pKa = 10.35HH92 pKa = 6.78NKK94 pKa = 8.49LTSFKK99 pKa = 10.56LWEE102 pKa = 4.12QGSNGTMALFPLQRR116 pKa = 11.84GTDD119 pKa = 3.16QDD121 pKa = 4.02EE122 pKa = 4.35IVGRR126 pKa = 11.84EE127 pKa = 3.99YY128 pKa = 10.86YY129 pKa = 10.77AKK131 pKa = 10.25SLKK134 pKa = 9.72IDD136 pKa = 3.55LQFTFPYY143 pKa = 10.26DD144 pKa = 3.51RR145 pKa = 11.84LGSTIKK151 pKa = 10.14VWYY154 pKa = 9.9VPVKK158 pKa = 9.89TGQPEE163 pKa = 3.64PAYY166 pKa = 10.71DD167 pKa = 3.37EE168 pKa = 4.77FFRR171 pKa = 11.84NTSGNIMLDD180 pKa = 3.27PRR182 pKa = 11.84NMQNYY187 pKa = 8.58PYY189 pKa = 10.6AKK191 pKa = 9.13YY192 pKa = 10.28LGSFRR197 pKa = 11.84PRR199 pKa = 11.84VKK201 pKa = 9.45QTTFGTLVNGNGVLGGRR218 pKa = 11.84DD219 pKa = 3.23ATVVKK224 pKa = 10.17SYY226 pKa = 10.61RR227 pKa = 11.84ISFNKK232 pKa = 9.41FFKK235 pKa = 10.62LRR237 pKa = 11.84SNVGGDD243 pKa = 3.64AEE245 pKa = 5.06IDD247 pKa = 3.86FNDD250 pKa = 3.94IPNLAEE256 pKa = 3.84KK257 pKa = 10.77AFLVFTAYY265 pKa = 10.63NEE267 pKa = 4.03EE268 pKa = 4.15AALEE272 pKa = 4.04TDD274 pKa = 3.54QVISNVEE281 pKa = 3.84GAFTFSYY288 pKa = 10.84KK289 pKa = 10.81DD290 pKa = 3.24PP291 pKa = 4.43
Molecular weight: 33.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
635 |
291 |
344 |
317.5 |
36.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.299 ± 0.8 | 0.787 ± 0.5 |
3.937 ± 0.118 | 7.717 ± 2.061 |
5.354 ± 0.527 | 6.142 ± 0.19 |
1.89 ± 0.981 | 3.937 ± 0.536 |
8.819 ± 0.728 | 5.354 ± 0.527 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.362 ± 0.464 | 5.354 ± 0.963 |
5.039 ± 0.145 | 4.882 ± 0.699 |
5.669 ± 0.328 | 5.669 ± 0.328 |
7.402 ± 0.755 | 5.984 ± 0.308 |
2.047 ± 0.645 | 5.354 ± 0.527 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
