
Microviridae sp. ctYqV29
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 8.27
Get precalculated fractions of proteins
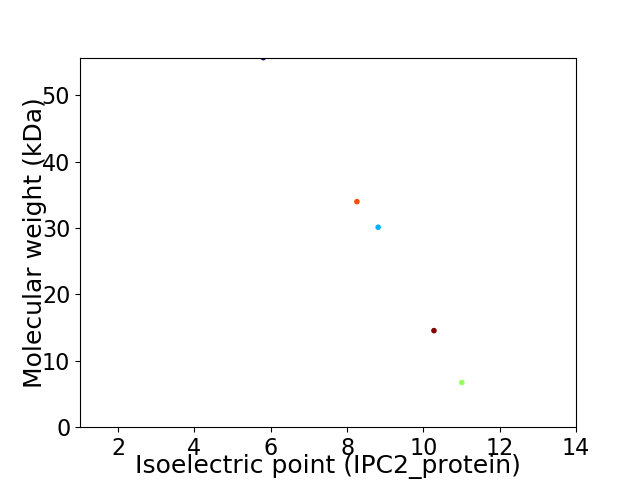
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W5J3|A0A5Q2W5J3_9VIRU Putative minor capsid protein OS=Microviridae sp. ctYqV29 OX=2656700 PE=4 SV=1
MM1 pKa = 7.25KK2 pKa = 10.06RR3 pKa = 11.84SKK5 pKa = 10.74FSLSNYY11 pKa = 9.7KK12 pKa = 10.33LLSCDD17 pKa = 2.93MGEE20 pKa = 4.67LVPCGLTEE28 pKa = 4.12VLPGDD33 pKa = 4.89TIQQATSCLLRR44 pKa = 11.84ASPLLSPVMHH54 pKa = 6.64PVDD57 pKa = 3.48VRR59 pKa = 11.84LHH61 pKa = 5.34HH62 pKa = 6.8WYY64 pKa = 10.27VPHH67 pKa = 6.81RR68 pKa = 11.84LVWEE72 pKa = 4.39DD73 pKa = 3.96FEE75 pKa = 6.02DD76 pKa = 5.54FITGGSDD83 pKa = 3.21GLDD86 pKa = 2.63ASVFPTITIGGGSGAAVGSLADD108 pKa = 3.82YY109 pKa = 10.83LGVPTGVNNIEE120 pKa = 4.13VSALPFRR127 pKa = 11.84GYY129 pKa = 11.66AMIWNEE135 pKa = 3.74WYY137 pKa = 10.17RR138 pKa = 11.84DD139 pKa = 3.38QDD141 pKa = 3.76LQTALAISEE150 pKa = 4.23GSGADD155 pKa = 3.43TTTSTVLQNCSWEE168 pKa = 3.99KK169 pKa = 11.18DD170 pKa = 3.58YY171 pKa = 9.31FTSSRR176 pKa = 11.84PWEE179 pKa = 4.19QKK181 pKa = 10.24GPAVTVPLGDD191 pKa = 3.58TAPVDD196 pKa = 4.56GISMRR201 pKa = 11.84NLGNAANNAGQSYY214 pKa = 8.86GAQAASDD221 pKa = 4.26YY222 pKa = 11.5GPGTTPTWRR231 pKa = 11.84ADD233 pKa = 3.53TQEE236 pKa = 4.17LYY238 pKa = 10.38IRR240 pKa = 11.84AQSNAVSGTGNRR252 pKa = 11.84PQIFADD258 pKa = 4.54LSNASAITVGALRR271 pKa = 11.84EE272 pKa = 4.07AMALQRR278 pKa = 11.84YY279 pKa = 8.41EE280 pKa = 3.73EE281 pKa = 3.96ARR283 pKa = 11.84ARR285 pKa = 11.84FGSRR289 pKa = 11.84YY290 pKa = 9.35VEE292 pKa = 3.84YY293 pKa = 10.7LRR295 pKa = 11.84YY296 pKa = 10.15LGVQSSDD303 pKa = 3.08ARR305 pKa = 11.84LQRR308 pKa = 11.84PEE310 pKa = 3.7YY311 pKa = 10.32LGGGRR316 pKa = 11.84EE317 pKa = 4.1TIQWSEE323 pKa = 3.99VLQTAEE329 pKa = 3.95GTDD332 pKa = 3.69PVGSLKK338 pKa = 10.9GHH340 pKa = 7.35GISSMRR346 pKa = 11.84SNRR349 pKa = 11.84YY350 pKa = 8.17RR351 pKa = 11.84RR352 pKa = 11.84YY353 pKa = 9.24IEE355 pKa = 3.61EE356 pKa = 3.87HH357 pKa = 6.26GYY359 pKa = 10.9VYY361 pKa = 10.58TFLSVRR367 pKa = 11.84PKK369 pKa = 10.02TIYY372 pKa = 10.16AQGLARR378 pKa = 11.84HH379 pKa = 5.55WNRR382 pKa = 11.84RR383 pKa = 11.84VKK385 pKa = 10.65EE386 pKa = 4.15DD387 pKa = 3.37FWQKK391 pKa = 9.59EE392 pKa = 4.02LQFIGQQEE400 pKa = 4.33VLNKK404 pKa = 9.76EE405 pKa = 4.16VYY407 pKa = 9.55AAHH410 pKa = 7.33ASPNNVFGYY419 pKa = 8.45QDD421 pKa = 3.44RR422 pKa = 11.84YY423 pKa = 10.92DD424 pKa = 3.54EE425 pKa = 4.09YY426 pKa = 11.35RR427 pKa = 11.84RR428 pKa = 11.84AEE430 pKa = 3.91STIAGEE436 pKa = 4.13YY437 pKa = 10.28RR438 pKa = 11.84SVLDD442 pKa = 4.13FWHH445 pKa = 6.73MARR448 pKa = 11.84IFGSSPALNGDD459 pKa = 3.57FVKK462 pKa = 10.67CVPTEE467 pKa = 3.71RR468 pKa = 11.84TFAVPSEE475 pKa = 4.05DD476 pKa = 4.72VLWCMVKK483 pKa = 10.44HH484 pKa = 6.47SIQARR489 pKa = 11.84RR490 pKa = 11.84LVAGVGKK497 pKa = 10.41SFIYY501 pKa = 10.53
MM1 pKa = 7.25KK2 pKa = 10.06RR3 pKa = 11.84SKK5 pKa = 10.74FSLSNYY11 pKa = 9.7KK12 pKa = 10.33LLSCDD17 pKa = 2.93MGEE20 pKa = 4.67LVPCGLTEE28 pKa = 4.12VLPGDD33 pKa = 4.89TIQQATSCLLRR44 pKa = 11.84ASPLLSPVMHH54 pKa = 6.64PVDD57 pKa = 3.48VRR59 pKa = 11.84LHH61 pKa = 5.34HH62 pKa = 6.8WYY64 pKa = 10.27VPHH67 pKa = 6.81RR68 pKa = 11.84LVWEE72 pKa = 4.39DD73 pKa = 3.96FEE75 pKa = 6.02DD76 pKa = 5.54FITGGSDD83 pKa = 3.21GLDD86 pKa = 2.63ASVFPTITIGGGSGAAVGSLADD108 pKa = 3.82YY109 pKa = 10.83LGVPTGVNNIEE120 pKa = 4.13VSALPFRR127 pKa = 11.84GYY129 pKa = 11.66AMIWNEE135 pKa = 3.74WYY137 pKa = 10.17RR138 pKa = 11.84DD139 pKa = 3.38QDD141 pKa = 3.76LQTALAISEE150 pKa = 4.23GSGADD155 pKa = 3.43TTTSTVLQNCSWEE168 pKa = 3.99KK169 pKa = 11.18DD170 pKa = 3.58YY171 pKa = 9.31FTSSRR176 pKa = 11.84PWEE179 pKa = 4.19QKK181 pKa = 10.24GPAVTVPLGDD191 pKa = 3.58TAPVDD196 pKa = 4.56GISMRR201 pKa = 11.84NLGNAANNAGQSYY214 pKa = 8.86GAQAASDD221 pKa = 4.26YY222 pKa = 11.5GPGTTPTWRR231 pKa = 11.84ADD233 pKa = 3.53TQEE236 pKa = 4.17LYY238 pKa = 10.38IRR240 pKa = 11.84AQSNAVSGTGNRR252 pKa = 11.84PQIFADD258 pKa = 4.54LSNASAITVGALRR271 pKa = 11.84EE272 pKa = 4.07AMALQRR278 pKa = 11.84YY279 pKa = 8.41EE280 pKa = 3.73EE281 pKa = 3.96ARR283 pKa = 11.84ARR285 pKa = 11.84FGSRR289 pKa = 11.84YY290 pKa = 9.35VEE292 pKa = 3.84YY293 pKa = 10.7LRR295 pKa = 11.84YY296 pKa = 10.15LGVQSSDD303 pKa = 3.08ARR305 pKa = 11.84LQRR308 pKa = 11.84PEE310 pKa = 3.7YY311 pKa = 10.32LGGGRR316 pKa = 11.84EE317 pKa = 4.1TIQWSEE323 pKa = 3.99VLQTAEE329 pKa = 3.95GTDD332 pKa = 3.69PVGSLKK338 pKa = 10.9GHH340 pKa = 7.35GISSMRR346 pKa = 11.84SNRR349 pKa = 11.84YY350 pKa = 8.17RR351 pKa = 11.84RR352 pKa = 11.84YY353 pKa = 9.24IEE355 pKa = 3.61EE356 pKa = 3.87HH357 pKa = 6.26GYY359 pKa = 10.9VYY361 pKa = 10.58TFLSVRR367 pKa = 11.84PKK369 pKa = 10.02TIYY372 pKa = 10.16AQGLARR378 pKa = 11.84HH379 pKa = 5.55WNRR382 pKa = 11.84RR383 pKa = 11.84VKK385 pKa = 10.65EE386 pKa = 4.15DD387 pKa = 3.37FWQKK391 pKa = 9.59EE392 pKa = 4.02LQFIGQQEE400 pKa = 4.33VLNKK404 pKa = 9.76EE405 pKa = 4.16VYY407 pKa = 9.55AAHH410 pKa = 7.33ASPNNVFGYY419 pKa = 8.45QDD421 pKa = 3.44RR422 pKa = 11.84YY423 pKa = 10.92DD424 pKa = 3.54EE425 pKa = 4.09YY426 pKa = 11.35RR427 pKa = 11.84RR428 pKa = 11.84AEE430 pKa = 3.91STIAGEE436 pKa = 4.13YY437 pKa = 10.28RR438 pKa = 11.84SVLDD442 pKa = 4.13FWHH445 pKa = 6.73MARR448 pKa = 11.84IFGSSPALNGDD459 pKa = 3.57FVKK462 pKa = 10.67CVPTEE467 pKa = 3.71RR468 pKa = 11.84TFAVPSEE475 pKa = 4.05DD476 pKa = 4.72VLWCMVKK483 pKa = 10.44HH484 pKa = 6.47SIQARR489 pKa = 11.84RR490 pKa = 11.84LVAGVGKK497 pKa = 10.41SFIYY501 pKa = 10.53
Molecular weight: 55.68 kDa
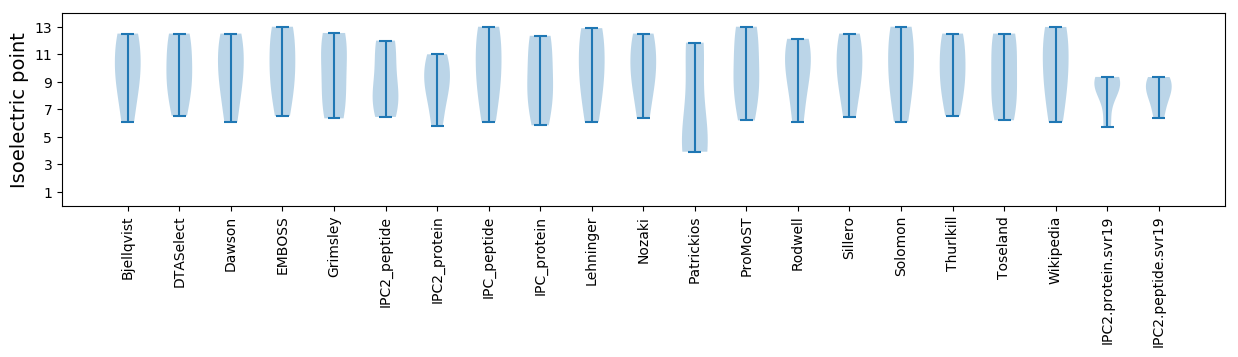
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W789|A0A5Q2W789_9VIRU Uncharacterized protein OS=Microviridae sp. ctYqV29 OX=2656700 PE=4 SV=1
MM1 pKa = 7.08VRR3 pKa = 11.84LGRR6 pKa = 11.84YY7 pKa = 7.71GWIIGGSEE15 pKa = 3.67MRR17 pKa = 11.84RR18 pKa = 11.84GFKK21 pKa = 10.17KK22 pKa = 10.27RR23 pKa = 11.84FARR26 pKa = 11.84GHH28 pKa = 5.15KK29 pKa = 9.49RR30 pKa = 11.84RR31 pKa = 11.84GRR33 pKa = 11.84GVRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84SNYY41 pKa = 7.16QRR43 pKa = 11.84RR44 pKa = 11.84KK45 pKa = 7.33GARR48 pKa = 11.84GVRR51 pKa = 11.84IGFRR55 pKa = 11.84MM56 pKa = 3.56
MM1 pKa = 7.08VRR3 pKa = 11.84LGRR6 pKa = 11.84YY7 pKa = 7.71GWIIGGSEE15 pKa = 3.67MRR17 pKa = 11.84RR18 pKa = 11.84GFKK21 pKa = 10.17KK22 pKa = 10.27RR23 pKa = 11.84FARR26 pKa = 11.84GHH28 pKa = 5.15KK29 pKa = 9.49RR30 pKa = 11.84RR31 pKa = 11.84GRR33 pKa = 11.84GVRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84SNYY41 pKa = 7.16QRR43 pKa = 11.84RR44 pKa = 11.84KK45 pKa = 7.33GARR48 pKa = 11.84GVRR51 pKa = 11.84IGFRR55 pKa = 11.84MM56 pKa = 3.56
Molecular weight: 6.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1260 |
56 |
501 |
252.0 |
28.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.492 ± 0.868 | 1.984 ± 1.172 |
5.079 ± 0.307 | 5.079 ± 0.865 |
2.937 ± 0.457 | 8.73 ± 0.885 |
1.508 ± 0.4 | 3.651 ± 0.298 |
4.841 ± 1.105 | 8.413 ± 0.669 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.222 ± 0.271 | 2.937 ± 0.381 |
4.841 ± 0.718 | 4.048 ± 0.693 |
10.0 ± 2.03 | 7.54 ± 0.742 |
4.762 ± 0.686 | 7.063 ± 0.772 |
1.905 ± 0.365 | 3.968 ± 0.565 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
