
Leptospirillum ferriphilum (strain ML-04)
Taxonomy: cellular organisms; Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Leptospirillum; Leptospirillum sp. Group III; Leptospirillum ferriphilum
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
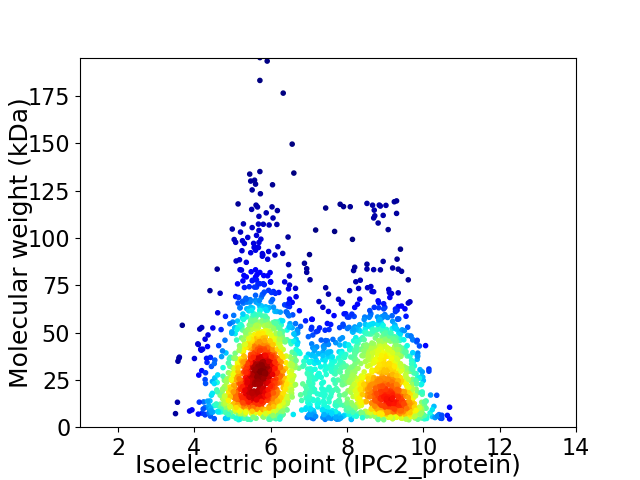
Virtual 2D-PAGE plot for 2450 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
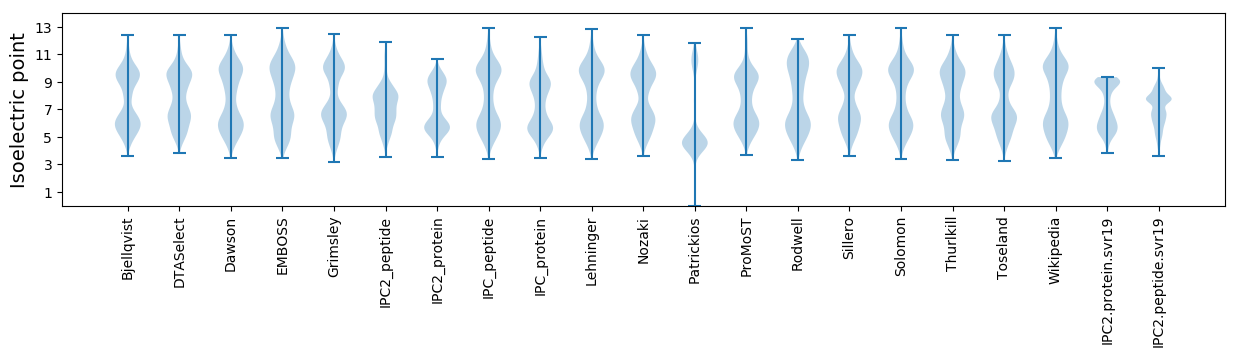
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9ZDA2|J9ZDA2_LEPFM Adenine phosphoribosyltransferase OS=Leptospirillum ferriphilum (strain ML-04) OX=1048260 GN=apt PE=3 SV=1
MM1 pKa = 7.6HH2 pKa = 7.04EE3 pKa = 4.1TFKK6 pKa = 11.18FLFSCRR12 pKa = 11.84YY13 pKa = 9.76ASVLLSLFALLASGGCNGDD32 pKa = 3.58NLDD35 pKa = 4.45FPSQGGPSATLLQGVAFDD53 pKa = 4.69GPILQGTVSLEE64 pKa = 3.83QYY66 pKa = 10.0PPGSGVLASGTTDD79 pKa = 3.11QNGNFSFTAPMLDD92 pKa = 3.22STAVYY97 pKa = 10.55VLTVSSGKK105 pKa = 8.2TLDD108 pKa = 3.74LATGNTLNLTQGDD121 pKa = 3.8QLMAIGTGQDD131 pKa = 3.43FSGGSLSITPFTSLEE146 pKa = 3.86ASLAEE151 pKa = 3.95HH152 pKa = 7.46FINQGIAMSDD162 pKa = 3.56ALRR165 pKa = 11.84QSDD168 pKa = 4.53DD169 pKa = 3.11LWSGYY174 pKa = 10.95LGFDD178 pKa = 3.68PLKK181 pKa = 10.21TPVANPTLGPTQANSAGLYY200 pKa = 10.24GIALAGLSQMAYY212 pKa = 10.7NIGQTQSLSPGTTNTFGLLIQLQSDD237 pKa = 4.11LSDD240 pKa = 4.37GIFNGLQQGSSTPLSYY256 pKa = 10.85YY257 pKa = 10.54GYY259 pKa = 10.49SLTSDD264 pKa = 3.59TLRR267 pKa = 11.84KK268 pKa = 9.73NLAAGILEE276 pKa = 4.35FLWNTQNKK284 pKa = 10.01SGLTPLTIDD293 pKa = 4.15GFANSLALNTSSLFSEE309 pKa = 4.96TPGASAPDD317 pKa = 3.85PSAPTISVVSPVVGQYY333 pKa = 10.79YY334 pKa = 10.4RR335 pKa = 11.84GTLTIMASASDD346 pKa = 3.83DD347 pKa = 3.84LGIGSFLTTSPNLPLSGGSLDD368 pKa = 3.96QNPLTTSFNTTSVADD383 pKa = 3.91GSYY386 pKa = 11.1NLVFTATDD394 pKa = 3.95YY395 pKa = 11.5AGHH398 pKa = 6.5TASQDD403 pKa = 3.4VQFSVDD409 pKa = 3.61NTPPTISNLSPTNDD423 pKa = 3.12QTLTFCAGITEE434 pKa = 4.68SVTVTGTLTDD444 pKa = 3.6MGSGPNSVGVSEE456 pKa = 4.64IAPTATQLTSSFTISSTNSTTGSFQFTFTVPADD489 pKa = 3.49GCKK492 pKa = 10.03SVTYY496 pKa = 10.32SFSLTGYY503 pKa = 10.6DD504 pKa = 3.46NLFNSSTIPYY514 pKa = 10.12SLTIEE519 pKa = 4.13NN520 pKa = 3.9
MM1 pKa = 7.6HH2 pKa = 7.04EE3 pKa = 4.1TFKK6 pKa = 11.18FLFSCRR12 pKa = 11.84YY13 pKa = 9.76ASVLLSLFALLASGGCNGDD32 pKa = 3.58NLDD35 pKa = 4.45FPSQGGPSATLLQGVAFDD53 pKa = 4.69GPILQGTVSLEE64 pKa = 3.83QYY66 pKa = 10.0PPGSGVLASGTTDD79 pKa = 3.11QNGNFSFTAPMLDD92 pKa = 3.22STAVYY97 pKa = 10.55VLTVSSGKK105 pKa = 8.2TLDD108 pKa = 3.74LATGNTLNLTQGDD121 pKa = 3.8QLMAIGTGQDD131 pKa = 3.43FSGGSLSITPFTSLEE146 pKa = 3.86ASLAEE151 pKa = 3.95HH152 pKa = 7.46FINQGIAMSDD162 pKa = 3.56ALRR165 pKa = 11.84QSDD168 pKa = 4.53DD169 pKa = 3.11LWSGYY174 pKa = 10.95LGFDD178 pKa = 3.68PLKK181 pKa = 10.21TPVANPTLGPTQANSAGLYY200 pKa = 10.24GIALAGLSQMAYY212 pKa = 10.7NIGQTQSLSPGTTNTFGLLIQLQSDD237 pKa = 4.11LSDD240 pKa = 4.37GIFNGLQQGSSTPLSYY256 pKa = 10.85YY257 pKa = 10.54GYY259 pKa = 10.49SLTSDD264 pKa = 3.59TLRR267 pKa = 11.84KK268 pKa = 9.73NLAAGILEE276 pKa = 4.35FLWNTQNKK284 pKa = 10.01SGLTPLTIDD293 pKa = 4.15GFANSLALNTSSLFSEE309 pKa = 4.96TPGASAPDD317 pKa = 3.85PSAPTISVVSPVVGQYY333 pKa = 10.79YY334 pKa = 10.4RR335 pKa = 11.84GTLTIMASASDD346 pKa = 3.83DD347 pKa = 3.84LGIGSFLTTSPNLPLSGGSLDD368 pKa = 3.96QNPLTTSFNTTSVADD383 pKa = 3.91GSYY386 pKa = 11.1NLVFTATDD394 pKa = 3.95YY395 pKa = 11.5AGHH398 pKa = 6.5TASQDD403 pKa = 3.4VQFSVDD409 pKa = 3.61NTPPTISNLSPTNDD423 pKa = 3.12QTLTFCAGITEE434 pKa = 4.68SVTVTGTLTDD444 pKa = 3.6MGSGPNSVGVSEE456 pKa = 4.64IAPTATQLTSSFTISSTNSTTGSFQFTFTVPADD489 pKa = 3.49GCKK492 pKa = 10.03SVTYY496 pKa = 10.32SFSLTGYY503 pKa = 10.6DD504 pKa = 3.46NLFNSSTIPYY514 pKa = 10.12SLTIEE519 pKa = 4.13NN520 pKa = 3.9
Molecular weight: 53.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9Z7F9|J9Z7F9_LEPFM Flagellar biosynthetic protein FliQ OS=Leptospirillum ferriphilum (strain ML-04) OX=1048260 GN=fliQ PE=3 SV=1
MM1 pKa = 7.44RR2 pKa = 11.84HH3 pKa = 6.11APLLLITIYY12 pKa = 10.73RR13 pKa = 11.84SGQTPAPDD21 pKa = 3.5SNRR24 pKa = 11.84RR25 pKa = 11.84ASFFFSVCVRR35 pKa = 11.84FPGGVNDD42 pKa = 4.19NSSTPAQRR50 pKa = 11.84GQLVRR55 pKa = 11.84LDD57 pKa = 3.4RR58 pKa = 11.84KK59 pKa = 9.72GVQKK63 pKa = 10.41IVEE66 pKa = 4.34NGDD69 pKa = 3.53RR70 pKa = 11.84MHH72 pKa = 6.87LQQASLHH79 pKa = 6.95PDD81 pKa = 3.47TAWTSEE87 pKa = 3.84HH88 pKa = 6.23SSEE91 pKa = 4.37RR92 pKa = 11.84FCEE95 pKa = 4.26TEE97 pKa = 3.45PAFAVVVSPDD107 pKa = 2.97VGGIRR112 pKa = 11.84KK113 pKa = 9.16PEE115 pKa = 4.02TVRR118 pKa = 11.84KK119 pKa = 9.98VLFHH123 pKa = 4.76VRR125 pKa = 11.84KK126 pKa = 9.66RR127 pKa = 11.84RR128 pKa = 11.84AHH130 pKa = 5.94GEE132 pKa = 3.79QGRR135 pKa = 11.84SRR137 pKa = 11.84GGIAPPGLVLFSGDD151 pKa = 2.73HH152 pKa = 6.63RR153 pKa = 11.84YY154 pKa = 9.63
MM1 pKa = 7.44RR2 pKa = 11.84HH3 pKa = 6.11APLLLITIYY12 pKa = 10.73RR13 pKa = 11.84SGQTPAPDD21 pKa = 3.5SNRR24 pKa = 11.84RR25 pKa = 11.84ASFFFSVCVRR35 pKa = 11.84FPGGVNDD42 pKa = 4.19NSSTPAQRR50 pKa = 11.84GQLVRR55 pKa = 11.84LDD57 pKa = 3.4RR58 pKa = 11.84KK59 pKa = 9.72GVQKK63 pKa = 10.41IVEE66 pKa = 4.34NGDD69 pKa = 3.53RR70 pKa = 11.84MHH72 pKa = 6.87LQQASLHH79 pKa = 6.95PDD81 pKa = 3.47TAWTSEE87 pKa = 3.84HH88 pKa = 6.23SSEE91 pKa = 4.37RR92 pKa = 11.84FCEE95 pKa = 4.26TEE97 pKa = 3.45PAFAVVVSPDD107 pKa = 2.97VGGIRR112 pKa = 11.84KK113 pKa = 9.16PEE115 pKa = 4.02TVRR118 pKa = 11.84KK119 pKa = 9.98VLFHH123 pKa = 4.76VRR125 pKa = 11.84KK126 pKa = 9.66RR127 pKa = 11.84RR128 pKa = 11.84AHH130 pKa = 5.94GEE132 pKa = 3.79QGRR135 pKa = 11.84SRR137 pKa = 11.84GGIAPPGLVLFSGDD151 pKa = 2.73HH152 pKa = 6.63RR153 pKa = 11.84YY154 pKa = 9.63
Molecular weight: 17.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
716377 |
37 |
1718 |
292.4 |
32.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.355 ± 0.05 | 0.793 ± 0.018 |
4.969 ± 0.032 | 6.501 ± 0.061 |
4.555 ± 0.039 | 8.148 ± 0.047 |
2.226 ± 0.022 | 5.558 ± 0.04 |
4.467 ± 0.043 | 11.135 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.395 ± 0.025 | 2.896 ± 0.034 |
5.749 ± 0.035 | 3.041 ± 0.026 |
7.292 ± 0.052 | 7.2 ± 0.044 |
4.999 ± 0.032 | 7.022 ± 0.047 |
1.296 ± 0.022 | 2.404 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
