
Roseburia hominis (strain DSM 16839 / JCM 17582 / NCIMB 14029 / A2-183)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; Roseburia hominis
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
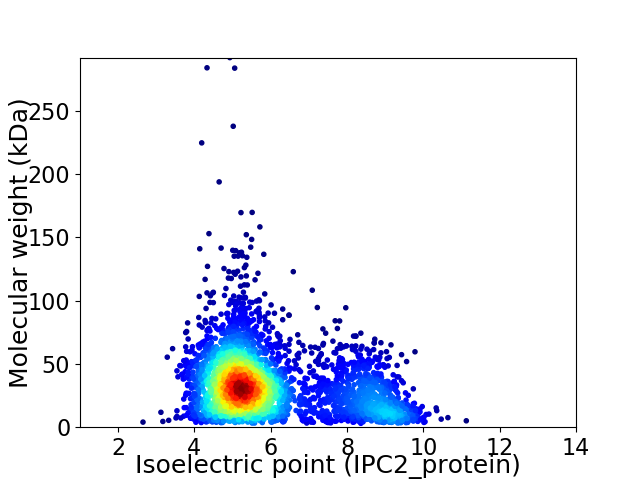
Virtual 2D-PAGE plot for 3351 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
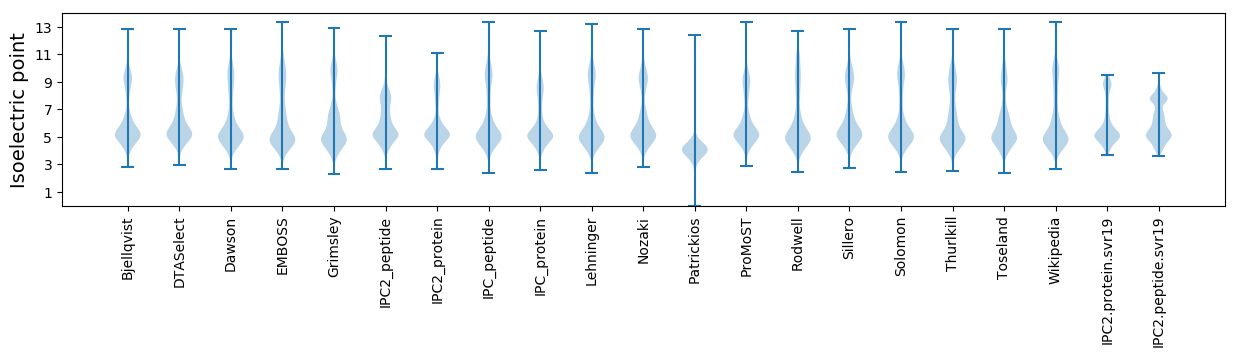
* You can choose from 21 different methods for calculating isoelectric point
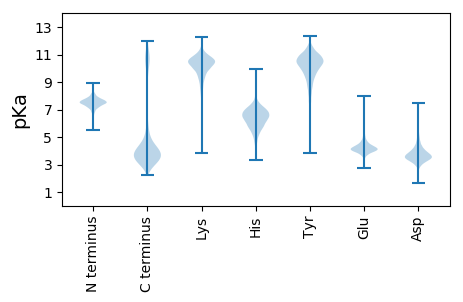
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2SY23|G2SY23_ROSHA ABC transporter (ATP-binding protein) OS=Roseburia hominis (strain DSM 16839 / JCM 17582 / NCIMB 14029 / A2-183) OX=585394 GN=RHOM_11705 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.06LKK4 pKa = 10.54KK5 pKa = 10.35VLAVSLAAAMTLSMAACGGSSSDD28 pKa = 3.87SSAASDD34 pKa = 3.68DD35 pKa = 3.65ATTGSVAATTTEE47 pKa = 4.0SGDD50 pKa = 3.6AAEE53 pKa = 4.56TTDD56 pKa = 3.73GALNYY61 pKa = 10.76ASIKK65 pKa = 10.5LGEE68 pKa = 5.17DD69 pKa = 3.3YY70 pKa = 11.32TDD72 pKa = 2.87ITTTIHH78 pKa = 6.17VFNQRR83 pKa = 11.84TDD85 pKa = 3.26MSEE88 pKa = 3.54DD89 pKa = 3.56SYY91 pKa = 11.57PGKK94 pKa = 10.31NWEE97 pKa = 4.48AYY99 pKa = 9.05IADD102 pKa = 4.22FNEE105 pKa = 4.38MYY107 pKa = 10.71PNITVEE113 pKa = 4.43VEE115 pKa = 3.44TDD117 pKa = 3.07TNYY120 pKa = 10.76SDD122 pKa = 6.0DD123 pKa = 3.7SLLRR127 pKa = 11.84LQSGDD132 pKa = 2.86WGDD135 pKa = 3.23IMMIPAVDD143 pKa = 3.5KK144 pKa = 11.32ADD146 pKa = 3.49LSEE149 pKa = 4.19YY150 pKa = 9.82FLPYY154 pKa = 9.0GTLDD158 pKa = 3.41EE159 pKa = 4.81MEE161 pKa = 4.39GQIKK165 pKa = 9.27FANTWDD171 pKa = 3.94YY172 pKa = 11.88DD173 pKa = 3.68GLVYY177 pKa = 10.31GVPSTGNAQGIVYY190 pKa = 9.49NKK192 pKa = 10.26RR193 pKa = 11.84VFTEE197 pKa = 4.0AGVAEE202 pKa = 4.62TPKK205 pKa = 10.7TPDD208 pKa = 2.91EE209 pKa = 5.0FIAALQAIKK218 pKa = 10.43DD219 pKa = 4.06YY220 pKa = 10.78DD221 pKa = 3.75SSIIPLYY228 pKa = 10.24TNYY231 pKa = 10.84ADD233 pKa = 3.82GWPMEE238 pKa = 4.09QWDD241 pKa = 4.14GQIAGTTTGDD251 pKa = 3.32STYY254 pKa = 10.5MNQKK258 pKa = 9.84LLHH261 pKa = 6.26TKK263 pKa = 10.61DD264 pKa = 3.36PFQDD268 pKa = 3.74YY269 pKa = 11.3GDD271 pKa = 3.78NTHH274 pKa = 7.41PYY276 pKa = 10.27ALYY279 pKa = 10.2KK280 pKa = 10.38VLYY283 pKa = 10.04DD284 pKa = 3.94AVADD288 pKa = 4.38GLTEE292 pKa = 5.36DD293 pKa = 5.66DD294 pKa = 4.06FTTTSWEE301 pKa = 4.14DD302 pKa = 3.16SKK304 pKa = 12.06GMINRR309 pKa = 11.84GEE311 pKa = 4.15IATMVLGSWAYY322 pKa = 10.91SQMVDD327 pKa = 3.46ADD329 pKa = 3.87SHH331 pKa = 7.73GEE333 pKa = 4.08DD334 pKa = 3.0IGYY337 pKa = 8.25MPFPITIDD345 pKa = 3.25GKK347 pKa = 10.34QYY349 pKa = 11.15ASAGADD355 pKa = 3.13YY356 pKa = 11.33SFGINAASDD365 pKa = 3.82ADD367 pKa = 3.85HH368 pKa = 6.37QAAAMVFVKK377 pKa = 9.95WMTEE381 pKa = 3.79EE382 pKa = 4.31SGFAYY387 pKa = 10.57NEE389 pKa = 3.9GGIPIAADD397 pKa = 3.57DD398 pKa = 4.15NDD400 pKa = 4.04YY401 pKa = 11.09PDD403 pKa = 4.76AYY405 pKa = 10.8AEE407 pKa = 4.28FGDD410 pKa = 3.76VTFVSDD416 pKa = 3.76EE417 pKa = 4.15SALEE421 pKa = 4.12GEE423 pKa = 4.37EE424 pKa = 4.94DD425 pKa = 5.06LLNALNADD433 pKa = 3.35SGLNINAGGKK443 pKa = 9.74EE444 pKa = 3.9KK445 pKa = 10.41VQEE448 pKa = 4.48IIEE451 pKa = 4.34HH452 pKa = 6.44ASNGDD457 pKa = 3.3MSYY460 pKa = 11.58DD461 pKa = 4.33DD462 pKa = 6.15IMAEE466 pKa = 4.17WNEE469 pKa = 4.0KK470 pKa = 6.58WTQAQEE476 pKa = 4.41DD477 pKa = 3.91NGVTAEE483 pKa = 3.93
MM1 pKa = 7.54KK2 pKa = 10.06LKK4 pKa = 10.54KK5 pKa = 10.35VLAVSLAAAMTLSMAACGGSSSDD28 pKa = 3.87SSAASDD34 pKa = 3.68DD35 pKa = 3.65ATTGSVAATTTEE47 pKa = 4.0SGDD50 pKa = 3.6AAEE53 pKa = 4.56TTDD56 pKa = 3.73GALNYY61 pKa = 10.76ASIKK65 pKa = 10.5LGEE68 pKa = 5.17DD69 pKa = 3.3YY70 pKa = 11.32TDD72 pKa = 2.87ITTTIHH78 pKa = 6.17VFNQRR83 pKa = 11.84TDD85 pKa = 3.26MSEE88 pKa = 3.54DD89 pKa = 3.56SYY91 pKa = 11.57PGKK94 pKa = 10.31NWEE97 pKa = 4.48AYY99 pKa = 9.05IADD102 pKa = 4.22FNEE105 pKa = 4.38MYY107 pKa = 10.71PNITVEE113 pKa = 4.43VEE115 pKa = 3.44TDD117 pKa = 3.07TNYY120 pKa = 10.76SDD122 pKa = 6.0DD123 pKa = 3.7SLLRR127 pKa = 11.84LQSGDD132 pKa = 2.86WGDD135 pKa = 3.23IMMIPAVDD143 pKa = 3.5KK144 pKa = 11.32ADD146 pKa = 3.49LSEE149 pKa = 4.19YY150 pKa = 9.82FLPYY154 pKa = 9.0GTLDD158 pKa = 3.41EE159 pKa = 4.81MEE161 pKa = 4.39GQIKK165 pKa = 9.27FANTWDD171 pKa = 3.94YY172 pKa = 11.88DD173 pKa = 3.68GLVYY177 pKa = 10.31GVPSTGNAQGIVYY190 pKa = 9.49NKK192 pKa = 10.26RR193 pKa = 11.84VFTEE197 pKa = 4.0AGVAEE202 pKa = 4.62TPKK205 pKa = 10.7TPDD208 pKa = 2.91EE209 pKa = 5.0FIAALQAIKK218 pKa = 10.43DD219 pKa = 4.06YY220 pKa = 10.78DD221 pKa = 3.75SSIIPLYY228 pKa = 10.24TNYY231 pKa = 10.84ADD233 pKa = 3.82GWPMEE238 pKa = 4.09QWDD241 pKa = 4.14GQIAGTTTGDD251 pKa = 3.32STYY254 pKa = 10.5MNQKK258 pKa = 9.84LLHH261 pKa = 6.26TKK263 pKa = 10.61DD264 pKa = 3.36PFQDD268 pKa = 3.74YY269 pKa = 11.3GDD271 pKa = 3.78NTHH274 pKa = 7.41PYY276 pKa = 10.27ALYY279 pKa = 10.2KK280 pKa = 10.38VLYY283 pKa = 10.04DD284 pKa = 3.94AVADD288 pKa = 4.38GLTEE292 pKa = 5.36DD293 pKa = 5.66DD294 pKa = 4.06FTTTSWEE301 pKa = 4.14DD302 pKa = 3.16SKK304 pKa = 12.06GMINRR309 pKa = 11.84GEE311 pKa = 4.15IATMVLGSWAYY322 pKa = 10.91SQMVDD327 pKa = 3.46ADD329 pKa = 3.87SHH331 pKa = 7.73GEE333 pKa = 4.08DD334 pKa = 3.0IGYY337 pKa = 8.25MPFPITIDD345 pKa = 3.25GKK347 pKa = 10.34QYY349 pKa = 11.15ASAGADD355 pKa = 3.13YY356 pKa = 11.33SFGINAASDD365 pKa = 3.82ADD367 pKa = 3.85HH368 pKa = 6.37QAAAMVFVKK377 pKa = 9.95WMTEE381 pKa = 3.79EE382 pKa = 4.31SGFAYY387 pKa = 10.57NEE389 pKa = 3.9GGIPIAADD397 pKa = 3.57DD398 pKa = 4.15NDD400 pKa = 4.04YY401 pKa = 11.09PDD403 pKa = 4.76AYY405 pKa = 10.8AEE407 pKa = 4.28FGDD410 pKa = 3.76VTFVSDD416 pKa = 3.76EE417 pKa = 4.15SALEE421 pKa = 4.12GEE423 pKa = 4.37EE424 pKa = 4.94DD425 pKa = 5.06LLNALNADD433 pKa = 3.35SGLNINAGGKK443 pKa = 9.74EE444 pKa = 3.9KK445 pKa = 10.41VQEE448 pKa = 4.48IIEE451 pKa = 4.34HH452 pKa = 6.44ASNGDD457 pKa = 3.3MSYY460 pKa = 11.58DD461 pKa = 4.33DD462 pKa = 6.15IMAEE466 pKa = 4.17WNEE469 pKa = 4.0KK470 pKa = 6.58WTQAQEE476 pKa = 4.41DD477 pKa = 3.91NGVTAEE483 pKa = 3.93
Molecular weight: 52.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2SWG8|G2SWG8_ROSHA Mutator family transposase OS=Roseburia hominis (strain DSM 16839 / JCM 17582 / NCIMB 14029 / A2-183) OX=585394 GN=RHOM_05665 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1052490 |
30 |
2665 |
314.1 |
35.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.25 ± 0.046 | 1.52 ± 0.018 |
5.732 ± 0.036 | 7.862 ± 0.052 |
3.903 ± 0.028 | 7.098 ± 0.039 |
1.822 ± 0.022 | 6.843 ± 0.041 |
6.17 ± 0.041 | 8.877 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.173 ± 0.024 | 4.119 ± 0.031 |
3.21 ± 0.025 | 3.441 ± 0.027 |
4.804 ± 0.038 | 5.515 ± 0.034 |
5.522 ± 0.041 | 6.981 ± 0.034 |
0.921 ± 0.014 | 4.236 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
