
Flavobacteriales bacterium
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; unclassified Flavobacteriales
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
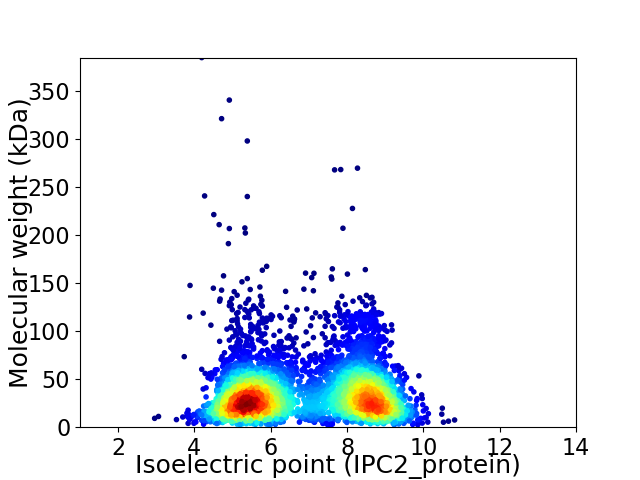
Virtual 2D-PAGE plot for 4745 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q3LXI3|A0A4Q3LXI3_9FLAO RagB/SusD family nutrient uptake outer membrane protein OS=Flavobacteriales bacterium OX=2021391 GN=EOO42_08670 PE=3 SV=1
MM1 pKa = 7.5KK2 pKa = 10.5NLMKK6 pKa = 10.17IVSCFLILSGIFARR20 pKa = 11.84AQADD24 pKa = 3.52PDD26 pKa = 4.23LEE28 pKa = 4.63ISTNSNTANGPAQSYY43 pKa = 7.7TLNFLKK49 pKa = 10.0DD50 pKa = 3.54TQNPTNVTAGPTPFVTAPNPLSVTLSISNSQYY82 pKa = 9.7ATAVTADD89 pKa = 4.27PLTQNALNLGYY100 pKa = 10.21SAGVPATTTLLSSGNGSVAANFTYY124 pKa = 10.2SGYY127 pKa = 10.85ANSAIGINPSGSNIASIRR145 pKa = 11.84NTGVQLMVATNQFSYY160 pKa = 11.25APYY163 pKa = 10.58KK164 pKa = 10.62KK165 pKa = 9.39LTNAKK170 pKa = 9.53IYY172 pKa = 10.35MGDD175 pKa = 3.23LTFTFSRR182 pKa = 11.84PVNNPVLHH190 pKa = 6.01FAGLGGSLGNQTLKK204 pKa = 10.39TSFEE208 pKa = 5.07LIGSAVALSRR218 pKa = 11.84LSGVPHH224 pKa = 6.97FDD226 pKa = 2.89VTSNVISSTSTGLLNTGNGNGSVLVTGRR254 pKa = 11.84GITSVSFKK262 pKa = 10.62IYY264 pKa = 9.31VTGDD268 pKa = 3.38GLNGADD274 pKa = 3.96ASTGYY279 pKa = 9.62TSTDD283 pKa = 2.43AWATNASGIFLPNNGTGGDD302 pKa = 3.9LMVISASLGEE312 pKa = 4.41SDD314 pKa = 5.26LSVTKK319 pKa = 10.01TVTNDD324 pKa = 3.29TPVVGSNVTFNIAATNSGMSKK345 pKa = 10.17SPEE348 pKa = 4.11TKK350 pKa = 9.47VTDD353 pKa = 4.37LLPSGFTYY361 pKa = 10.65VSSTTSVGTYY371 pKa = 10.64NSVTGIWDD379 pKa = 3.32IGEE382 pKa = 4.09MAIGAIATMSITATVKK398 pKa = 10.58SSGIHH403 pKa = 5.86TNTATIAGHH412 pKa = 5.48NTDD415 pKa = 4.72SNLNNNTASALVQQDD430 pKa = 3.0SDD432 pKa = 3.91GDD434 pKa = 4.27GVPDD438 pKa = 3.92TTDD441 pKa = 4.51LDD443 pKa = 4.3DD444 pKa = 6.11DD445 pKa = 4.44NDD447 pKa = 5.06GILDD451 pKa = 3.63SSEE454 pKa = 4.62NGPCTATSPLSSTGWKK470 pKa = 8.05ATVYY474 pKa = 9.59DD475 pKa = 4.55APSSTDD481 pKa = 2.53AWGEE485 pKa = 3.59ISAATTFPTASYY497 pKa = 10.68YY498 pKa = 10.28PIATFDD504 pKa = 3.7YY505 pKa = 11.05NEE507 pKa = 4.01FAGTSDD513 pKa = 3.96AFDD516 pKa = 4.35FNFATGAIGLKK527 pKa = 9.92AANPKK532 pKa = 7.28VTNYY536 pKa = 10.33VGGTIDD542 pKa = 3.7GNAFGSPVNTQDD554 pKa = 4.54DD555 pKa = 4.32YY556 pKa = 12.04AIMFRR561 pKa = 11.84KK562 pKa = 8.45TITAQEE568 pKa = 4.01AGTYY572 pKa = 10.69AFDD575 pKa = 4.27FLYY578 pKa = 11.0GDD580 pKa = 4.29DD581 pKa = 4.13QLFIYY586 pKa = 10.36KK587 pKa = 10.29NGVKK591 pKa = 10.19VYY593 pKa = 10.8QLLQAYY599 pKa = 7.51NVPATNNAATITVVEE614 pKa = 4.16GDD616 pKa = 4.23VISMLVAEE624 pKa = 4.49EE625 pKa = 4.33NTWNTEE631 pKa = 2.98IRR633 pKa = 11.84FNGRR637 pKa = 11.84KK638 pKa = 8.84IAGTCTLTDD647 pKa = 3.45TDD649 pKa = 4.53GDD651 pKa = 4.45GIPNYY656 pKa = 10.62LDD658 pKa = 4.76LDD660 pKa = 4.14SDD662 pKa = 4.4NDD664 pKa = 3.76GCTDD668 pKa = 3.52ARR670 pKa = 11.84EE671 pKa = 4.04GDD673 pKa = 4.04EE674 pKa = 4.99NVLASQLVTASGTLTVGTGSTASNQNLGITVDD706 pKa = 3.64ANGVPTLVNSGGAADD721 pKa = 4.08IGADD725 pKa = 3.34QGQGIGVSQDD735 pKa = 3.27SAKK738 pKa = 10.52SDD740 pKa = 3.97CLDD743 pKa = 3.41SDD745 pKa = 4.2GDD747 pKa = 4.44GVPDD751 pKa = 4.49WSDD754 pKa = 4.26LDD756 pKa = 4.45DD757 pKa = 5.87DD758 pKa = 4.93NDD760 pKa = 5.02GILDD764 pKa = 4.03TEE766 pKa = 4.58EE767 pKa = 4.61LRR769 pKa = 11.84CDD771 pKa = 3.99YY772 pKa = 10.93NPSVYY777 pKa = 10.81NFVSGSGAIRR787 pKa = 11.84NQLLIFDD794 pKa = 4.03WTGASLTNVNDD805 pKa = 3.58TFTTSKK811 pKa = 8.92TVNGITYY818 pKa = 7.21TAKK821 pKa = 9.23ATVVEE826 pKa = 4.41LVGGYY831 pKa = 10.05YY832 pKa = 10.19SGQSPIPSILNTFPGGGNQMFWRR855 pKa = 11.84YY856 pKa = 10.12ANYY859 pKa = 8.0GTTATDD865 pKa = 3.76DD866 pKa = 3.45LRR868 pKa = 11.84GAIVFQLARR877 pKa = 11.84VANNKK882 pKa = 9.92LSIKK886 pKa = 10.29FDD888 pKa = 3.01ITATKK893 pKa = 10.49GGLPQQFDD901 pKa = 4.53AIAFDD906 pKa = 4.34AEE908 pKa = 4.49STTTAEE914 pKa = 4.11KK915 pKa = 10.94GLYY918 pKa = 5.92TTNAADD924 pKa = 3.16WTLIDD929 pKa = 4.12NLGGVPTATEE939 pKa = 3.62ASIVGKK945 pKa = 7.36QLRR948 pKa = 11.84YY949 pKa = 10.53DD950 pKa = 3.78DD951 pKa = 4.16TQTAQLSLFAYY962 pKa = 8.13TRR964 pKa = 11.84GVNVSILGEE973 pKa = 4.09YY974 pKa = 10.71SNEE977 pKa = 3.95TNLARR982 pKa = 11.84AQAMGVGVYY991 pKa = 9.21LYY993 pKa = 10.83CDD995 pKa = 3.88ADD997 pKa = 3.55NDD999 pKa = 4.17GTFNYY1004 pKa = 10.53LDD1006 pKa = 4.84LDD1008 pKa = 4.1SDD1010 pKa = 4.42NDD1012 pKa = 3.86GCLDD1016 pKa = 3.57ALEE1019 pKa = 5.16GGSALAISNLVNASGTVSVGTGSTASNQNLCASSSCVGSTGIPTIVNQTSGQTGGSSTNNLVKK1082 pKa = 10.51AAVCSVCYY1090 pKa = 9.44EE1091 pKa = 4.29LPTDD1095 pKa = 4.27LTTSVPVSHH1104 pKa = 7.34GITVV1108 pKa = 3.5
MM1 pKa = 7.5KK2 pKa = 10.5NLMKK6 pKa = 10.17IVSCFLILSGIFARR20 pKa = 11.84AQADD24 pKa = 3.52PDD26 pKa = 4.23LEE28 pKa = 4.63ISTNSNTANGPAQSYY43 pKa = 7.7TLNFLKK49 pKa = 10.0DD50 pKa = 3.54TQNPTNVTAGPTPFVTAPNPLSVTLSISNSQYY82 pKa = 9.7ATAVTADD89 pKa = 4.27PLTQNALNLGYY100 pKa = 10.21SAGVPATTTLLSSGNGSVAANFTYY124 pKa = 10.2SGYY127 pKa = 10.85ANSAIGINPSGSNIASIRR145 pKa = 11.84NTGVQLMVATNQFSYY160 pKa = 11.25APYY163 pKa = 10.58KK164 pKa = 10.62KK165 pKa = 9.39LTNAKK170 pKa = 9.53IYY172 pKa = 10.35MGDD175 pKa = 3.23LTFTFSRR182 pKa = 11.84PVNNPVLHH190 pKa = 6.01FAGLGGSLGNQTLKK204 pKa = 10.39TSFEE208 pKa = 5.07LIGSAVALSRR218 pKa = 11.84LSGVPHH224 pKa = 6.97FDD226 pKa = 2.89VTSNVISSTSTGLLNTGNGNGSVLVTGRR254 pKa = 11.84GITSVSFKK262 pKa = 10.62IYY264 pKa = 9.31VTGDD268 pKa = 3.38GLNGADD274 pKa = 3.96ASTGYY279 pKa = 9.62TSTDD283 pKa = 2.43AWATNASGIFLPNNGTGGDD302 pKa = 3.9LMVISASLGEE312 pKa = 4.41SDD314 pKa = 5.26LSVTKK319 pKa = 10.01TVTNDD324 pKa = 3.29TPVVGSNVTFNIAATNSGMSKK345 pKa = 10.17SPEE348 pKa = 4.11TKK350 pKa = 9.47VTDD353 pKa = 4.37LLPSGFTYY361 pKa = 10.65VSSTTSVGTYY371 pKa = 10.64NSVTGIWDD379 pKa = 3.32IGEE382 pKa = 4.09MAIGAIATMSITATVKK398 pKa = 10.58SSGIHH403 pKa = 5.86TNTATIAGHH412 pKa = 5.48NTDD415 pKa = 4.72SNLNNNTASALVQQDD430 pKa = 3.0SDD432 pKa = 3.91GDD434 pKa = 4.27GVPDD438 pKa = 3.92TTDD441 pKa = 4.51LDD443 pKa = 4.3DD444 pKa = 6.11DD445 pKa = 4.44NDD447 pKa = 5.06GILDD451 pKa = 3.63SSEE454 pKa = 4.62NGPCTATSPLSSTGWKK470 pKa = 8.05ATVYY474 pKa = 9.59DD475 pKa = 4.55APSSTDD481 pKa = 2.53AWGEE485 pKa = 3.59ISAATTFPTASYY497 pKa = 10.68YY498 pKa = 10.28PIATFDD504 pKa = 3.7YY505 pKa = 11.05NEE507 pKa = 4.01FAGTSDD513 pKa = 3.96AFDD516 pKa = 4.35FNFATGAIGLKK527 pKa = 9.92AANPKK532 pKa = 7.28VTNYY536 pKa = 10.33VGGTIDD542 pKa = 3.7GNAFGSPVNTQDD554 pKa = 4.54DD555 pKa = 4.32YY556 pKa = 12.04AIMFRR561 pKa = 11.84KK562 pKa = 8.45TITAQEE568 pKa = 4.01AGTYY572 pKa = 10.69AFDD575 pKa = 4.27FLYY578 pKa = 11.0GDD580 pKa = 4.29DD581 pKa = 4.13QLFIYY586 pKa = 10.36KK587 pKa = 10.29NGVKK591 pKa = 10.19VYY593 pKa = 10.8QLLQAYY599 pKa = 7.51NVPATNNAATITVVEE614 pKa = 4.16GDD616 pKa = 4.23VISMLVAEE624 pKa = 4.49EE625 pKa = 4.33NTWNTEE631 pKa = 2.98IRR633 pKa = 11.84FNGRR637 pKa = 11.84KK638 pKa = 8.84IAGTCTLTDD647 pKa = 3.45TDD649 pKa = 4.53GDD651 pKa = 4.45GIPNYY656 pKa = 10.62LDD658 pKa = 4.76LDD660 pKa = 4.14SDD662 pKa = 4.4NDD664 pKa = 3.76GCTDD668 pKa = 3.52ARR670 pKa = 11.84EE671 pKa = 4.04GDD673 pKa = 4.04EE674 pKa = 4.99NVLASQLVTASGTLTVGTGSTASNQNLGITVDD706 pKa = 3.64ANGVPTLVNSGGAADD721 pKa = 4.08IGADD725 pKa = 3.34QGQGIGVSQDD735 pKa = 3.27SAKK738 pKa = 10.52SDD740 pKa = 3.97CLDD743 pKa = 3.41SDD745 pKa = 4.2GDD747 pKa = 4.44GVPDD751 pKa = 4.49WSDD754 pKa = 4.26LDD756 pKa = 4.45DD757 pKa = 5.87DD758 pKa = 4.93NDD760 pKa = 5.02GILDD764 pKa = 4.03TEE766 pKa = 4.58EE767 pKa = 4.61LRR769 pKa = 11.84CDD771 pKa = 3.99YY772 pKa = 10.93NPSVYY777 pKa = 10.81NFVSGSGAIRR787 pKa = 11.84NQLLIFDD794 pKa = 4.03WTGASLTNVNDD805 pKa = 3.58TFTTSKK811 pKa = 8.92TVNGITYY818 pKa = 7.21TAKK821 pKa = 9.23ATVVEE826 pKa = 4.41LVGGYY831 pKa = 10.05YY832 pKa = 10.19SGQSPIPSILNTFPGGGNQMFWRR855 pKa = 11.84YY856 pKa = 10.12ANYY859 pKa = 8.0GTTATDD865 pKa = 3.76DD866 pKa = 3.45LRR868 pKa = 11.84GAIVFQLARR877 pKa = 11.84VANNKK882 pKa = 9.92LSIKK886 pKa = 10.29FDD888 pKa = 3.01ITATKK893 pKa = 10.49GGLPQQFDD901 pKa = 4.53AIAFDD906 pKa = 4.34AEE908 pKa = 4.49STTTAEE914 pKa = 4.11KK915 pKa = 10.94GLYY918 pKa = 5.92TTNAADD924 pKa = 3.16WTLIDD929 pKa = 4.12NLGGVPTATEE939 pKa = 3.62ASIVGKK945 pKa = 7.36QLRR948 pKa = 11.84YY949 pKa = 10.53DD950 pKa = 3.78DD951 pKa = 4.16TQTAQLSLFAYY962 pKa = 8.13TRR964 pKa = 11.84GVNVSILGEE973 pKa = 4.09YY974 pKa = 10.71SNEE977 pKa = 3.95TNLARR982 pKa = 11.84AQAMGVGVYY991 pKa = 9.21LYY993 pKa = 10.83CDD995 pKa = 3.88ADD997 pKa = 3.55NDD999 pKa = 4.17GTFNYY1004 pKa = 10.53LDD1006 pKa = 4.84LDD1008 pKa = 4.1SDD1010 pKa = 4.42NDD1012 pKa = 3.86GCLDD1016 pKa = 3.57ALEE1019 pKa = 5.16GGSALAISNLVNASGTVSVGTGSTASNQNLCASSSCVGSTGIPTIVNQTSGQTGGSSTNNLVKK1082 pKa = 10.51AAVCSVCYY1090 pKa = 9.44EE1091 pKa = 4.29LPTDD1095 pKa = 4.27LTTSVPVSHH1104 pKa = 7.34GITVV1108 pKa = 3.5
Molecular weight: 114.82 kDa
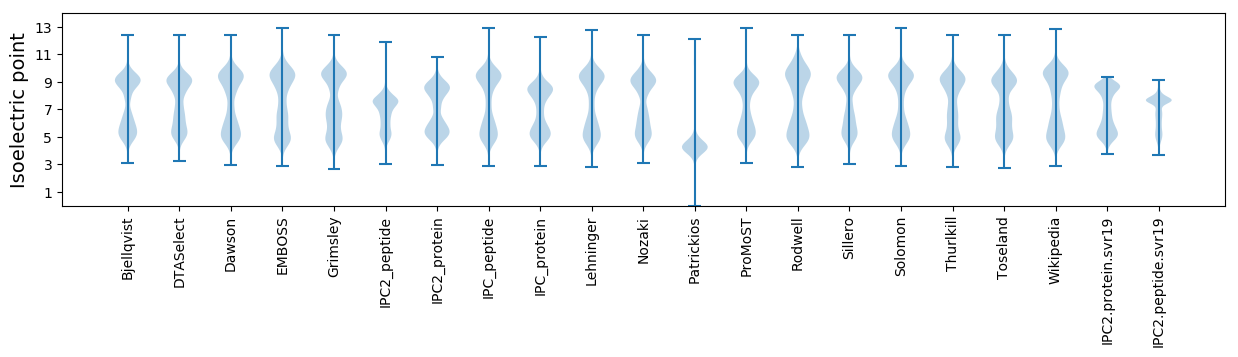
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q3LPY7|A0A4Q3LPY7_9FLAO Malonyl CoA-acyl carrier protein transacylase OS=Flavobacteriales bacterium OX=2021391 GN=fabD PE=3 SV=1
MM1 pKa = 7.32NALSPTKK8 pKa = 10.14TIMRR12 pKa = 11.84TDD14 pKa = 2.9ITLTTEE20 pKa = 3.91RR21 pKa = 11.84KK22 pKa = 7.2TPYY25 pKa = 8.81NTGFASGGLTCKK37 pKa = 10.63LGALCFYY44 pKa = 10.27LSSVLADD51 pKa = 3.45SFVLRR56 pKa = 11.84SPPEE60 pKa = 3.56RR61 pKa = 11.84KK62 pKa = 9.37ARR64 pKa = 11.84NRR66 pKa = 11.84WRR68 pKa = 3.48
MM1 pKa = 7.32NALSPTKK8 pKa = 10.14TIMRR12 pKa = 11.84TDD14 pKa = 2.9ITLTTEE20 pKa = 3.91RR21 pKa = 11.84KK22 pKa = 7.2TPYY25 pKa = 8.81NTGFASGGLTCKK37 pKa = 10.63LGALCFYY44 pKa = 10.27LSSVLADD51 pKa = 3.45SFVLRR56 pKa = 11.84SPPEE60 pKa = 3.56RR61 pKa = 11.84KK62 pKa = 9.37ARR64 pKa = 11.84NRR66 pKa = 11.84WRR68 pKa = 3.48
Molecular weight: 7.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1671011 |
22 |
3715 |
352.2 |
39.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.721 ± 0.038 | 0.679 ± 0.012 |
5.251 ± 0.023 | 5.144 ± 0.037 |
4.834 ± 0.027 | 7.233 ± 0.037 |
1.453 ± 0.02 | 7.071 ± 0.032 |
6.844 ± 0.046 | 9.054 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.981 ± 0.02 | 6.582 ± 0.043 |
3.678 ± 0.02 | 3.603 ± 0.021 |
3.594 ± 0.024 | 6.701 ± 0.034 |
6.59 ± 0.063 | 6.594 ± 0.033 |
1.122 ± 0.014 | 4.27 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
