
Prunus geminivirus A
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Grablovirus; Prunus latent virus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
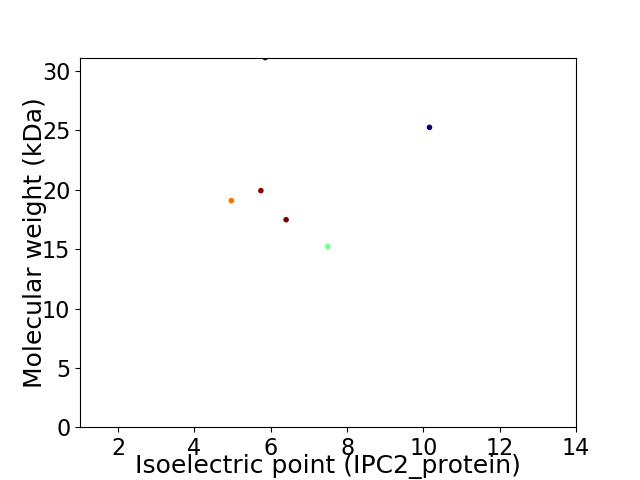
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
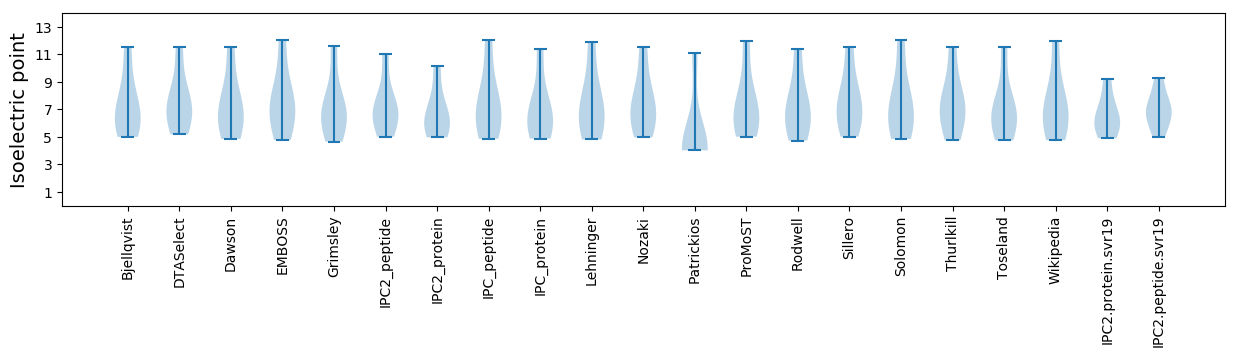
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L1GUQ4|A0A2L1GUQ4_9GEMI C3 Protein OS=Prunus geminivirus A OX=2022321 PE=4 SV=1
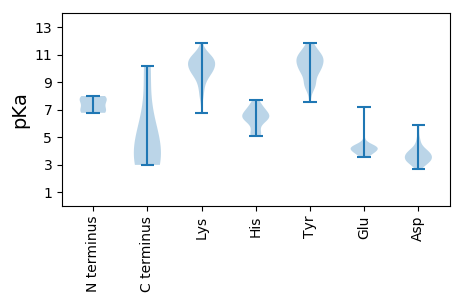
MM1 pKa = 7.11MFLVVVGMMLSVIAYY16 pKa = 7.56IWRR19 pKa = 11.84VLLLPDD25 pKa = 3.81ILSGYY30 pKa = 10.05ILMGLLISIMVLFFHH45 pKa = 7.7RR46 pKa = 11.84PRR48 pKa = 11.84CLIWRR53 pKa = 11.84VYY55 pKa = 10.71DD56 pKa = 3.48QVSEE60 pKa = 4.09VGLYY64 pKa = 10.39DD65 pKa = 4.58GYY67 pKa = 11.54DD68 pKa = 3.53DD69 pKa = 4.03VVKK72 pKa = 10.54DD73 pKa = 3.81IPIDD77 pKa = 3.98GDD79 pKa = 3.65DD80 pKa = 4.36DD81 pKa = 4.08EE82 pKa = 7.19SYY84 pKa = 10.26MVSDD88 pKa = 4.91EE89 pKa = 4.09YY90 pKa = 11.85AEE92 pKa = 3.95FLEE95 pKa = 4.26TLKK98 pKa = 10.55RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84SEE103 pKa = 4.0RR104 pKa = 11.84GGMTSNLKK112 pKa = 9.88RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84VMVDD119 pKa = 2.91PDD121 pKa = 3.44EE122 pKa = 5.79DD123 pKa = 3.68LVNRR127 pKa = 11.84LKK129 pKa = 10.76EE130 pKa = 3.95YY131 pKa = 10.63LGEE134 pKa = 4.34DD135 pKa = 3.32EE136 pKa = 4.35YY137 pKa = 11.76DD138 pKa = 2.99RR139 pKa = 11.84HH140 pKa = 6.56RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84LDD145 pKa = 3.28LEE147 pKa = 4.68LGSGQHH153 pKa = 6.27RR154 pKa = 11.84PDD156 pKa = 3.31SRR158 pKa = 11.84CVVMM162 pKa = 5.93
MM1 pKa = 7.11MFLVVVGMMLSVIAYY16 pKa = 7.56IWRR19 pKa = 11.84VLLLPDD25 pKa = 3.81ILSGYY30 pKa = 10.05ILMGLLISIMVLFFHH45 pKa = 7.7RR46 pKa = 11.84PRR48 pKa = 11.84CLIWRR53 pKa = 11.84VYY55 pKa = 10.71DD56 pKa = 3.48QVSEE60 pKa = 4.09VGLYY64 pKa = 10.39DD65 pKa = 4.58GYY67 pKa = 11.54DD68 pKa = 3.53DD69 pKa = 4.03VVKK72 pKa = 10.54DD73 pKa = 3.81IPIDD77 pKa = 3.98GDD79 pKa = 3.65DD80 pKa = 4.36DD81 pKa = 4.08EE82 pKa = 7.19SYY84 pKa = 10.26MVSDD88 pKa = 4.91EE89 pKa = 4.09YY90 pKa = 11.85AEE92 pKa = 3.95FLEE95 pKa = 4.26TLKK98 pKa = 10.55RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84SEE103 pKa = 4.0RR104 pKa = 11.84GGMTSNLKK112 pKa = 9.88RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84VMVDD119 pKa = 2.91PDD121 pKa = 3.44EE122 pKa = 5.79DD123 pKa = 3.68LVNRR127 pKa = 11.84LKK129 pKa = 10.76EE130 pKa = 3.95YY131 pKa = 10.63LGEE134 pKa = 4.34DD135 pKa = 3.32EE136 pKa = 4.35YY137 pKa = 11.76DD138 pKa = 2.99RR139 pKa = 11.84HH140 pKa = 6.56RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84LDD145 pKa = 3.28LEE147 pKa = 4.68LGSGQHH153 pKa = 6.27RR154 pKa = 11.84PDD156 pKa = 3.31SRR158 pKa = 11.84CVVMM162 pKa = 5.93
Molecular weight: 19.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L1GUS0|A0A2L1GUS0_9GEMI CP/V1 protein OS=Prunus geminivirus A OX=2022321 GN=CP PE=4 SV=1
MM1 pKa = 6.78VLKK4 pKa = 10.39RR5 pKa = 11.84RR6 pKa = 11.84PSRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 9.24RR12 pKa = 11.84RR13 pKa = 11.84ATKK16 pKa = 9.95KK17 pKa = 10.24RR18 pKa = 11.84SGRR21 pKa = 11.84SSQRR25 pKa = 11.84KK26 pKa = 7.87RR27 pKa = 11.84RR28 pKa = 11.84ARR30 pKa = 11.84PRR32 pKa = 11.84SRR34 pKa = 11.84PCQFSFHH41 pKa = 6.31GNAFGGTPTLFFLTPIALGNGAEE64 pKa = 4.26DD65 pKa = 3.4RR66 pKa = 11.84TGSVLTVSSMYY77 pKa = 10.75LKK79 pKa = 10.74GVVVPTDD86 pKa = 3.17NVTDD90 pKa = 4.27GLHH93 pKa = 7.49DD94 pKa = 3.64IYY96 pKa = 11.1FWIILDD102 pKa = 4.21RR103 pKa = 11.84FPTGTDD109 pKa = 3.28PAVSDD114 pKa = 3.93IFTGSDD120 pKa = 2.79TGGSMIEE127 pKa = 3.9TLTRR131 pKa = 11.84NRR133 pKa = 11.84LNRR136 pKa = 11.84KK137 pKa = 8.14RR138 pKa = 11.84FRR140 pKa = 11.84ILGSKK145 pKa = 10.12KK146 pKa = 10.37LVIGVNRR153 pKa = 11.84KK154 pKa = 7.78PQEE157 pKa = 4.33SLPHH161 pKa = 5.12TRR163 pKa = 11.84AAFNIFQRR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84LVVAFKK179 pKa = 11.03NDD181 pKa = 3.04VSGGGRR187 pKa = 11.84NDD189 pKa = 3.11VEE191 pKa = 4.26RR192 pKa = 11.84NRR194 pKa = 11.84IYY196 pKa = 9.69MACASATGHH205 pKa = 6.1SFRR208 pKa = 11.84LYY210 pKa = 11.09LNGIVNFYY218 pKa = 10.77NGVIFSS224 pKa = 3.89
MM1 pKa = 6.78VLKK4 pKa = 10.39RR5 pKa = 11.84RR6 pKa = 11.84PSRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 9.24RR12 pKa = 11.84RR13 pKa = 11.84ATKK16 pKa = 9.95KK17 pKa = 10.24RR18 pKa = 11.84SGRR21 pKa = 11.84SSQRR25 pKa = 11.84KK26 pKa = 7.87RR27 pKa = 11.84RR28 pKa = 11.84ARR30 pKa = 11.84PRR32 pKa = 11.84SRR34 pKa = 11.84PCQFSFHH41 pKa = 6.31GNAFGGTPTLFFLTPIALGNGAEE64 pKa = 4.26DD65 pKa = 3.4RR66 pKa = 11.84TGSVLTVSSMYY77 pKa = 10.75LKK79 pKa = 10.74GVVVPTDD86 pKa = 3.17NVTDD90 pKa = 4.27GLHH93 pKa = 7.49DD94 pKa = 3.64IYY96 pKa = 11.1FWIILDD102 pKa = 4.21RR103 pKa = 11.84FPTGTDD109 pKa = 3.28PAVSDD114 pKa = 3.93IFTGSDD120 pKa = 2.79TGGSMIEE127 pKa = 3.9TLTRR131 pKa = 11.84NRR133 pKa = 11.84LNRR136 pKa = 11.84KK137 pKa = 8.14RR138 pKa = 11.84FRR140 pKa = 11.84ILGSKK145 pKa = 10.12KK146 pKa = 10.37LVIGVNRR153 pKa = 11.84KK154 pKa = 7.78PQEE157 pKa = 4.33SLPHH161 pKa = 5.12TRR163 pKa = 11.84AAFNIFQRR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84LVVAFKK179 pKa = 11.03NDD181 pKa = 3.04VSGGGRR187 pKa = 11.84NDD189 pKa = 3.11VEE191 pKa = 4.26RR192 pKa = 11.84NRR194 pKa = 11.84IYY196 pKa = 9.69MACASATGHH205 pKa = 6.1SFRR208 pKa = 11.84LYY210 pKa = 11.09LNGIVNFYY218 pKa = 10.77NGVIFSS224 pKa = 3.89
Molecular weight: 25.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1113 |
131 |
262 |
185.5 |
21.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.953 ± 0.872 | 1.617 ± 0.472 |
6.11 ± 1.121 | 5.75 ± 0.988 |
4.403 ± 0.747 | 5.211 ± 1.168 |
2.695 ± 0.631 | 5.481 ± 0.523 |
5.301 ± 0.914 | 7.907 ± 0.922 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.516 ± 0.688 | 4.672 ± 0.877 |
4.942 ± 0.417 | 3.684 ± 0.809 |
7.817 ± 1.805 | 9.344 ± 2.013 |
6.739 ± 1.636 | 6.289 ± 1.247 |
1.527 ± 0.431 | 4.043 ± 0.633 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
