
Paracoccus homiensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
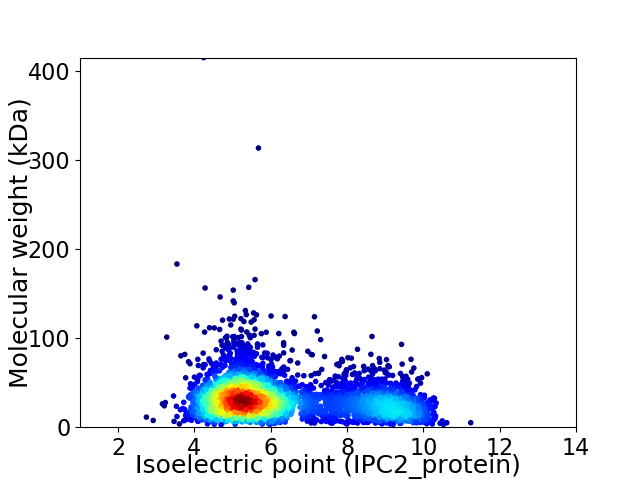
Virtual 2D-PAGE plot for 3828 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9ZXL6|A0A1H9ZXL6_9RHOB Nucleoside-binding protein OS=Paracoccus homiensis OX=364199 GN=SAMN04489858_10230 PE=4 SV=1
MM1 pKa = 7.33ATLFGNSGNNTIPGTSGADD20 pKa = 3.78LIYY23 pKa = 10.85GFGGNDD29 pKa = 3.16SLGGNGGRR37 pKa = 11.84DD38 pKa = 4.01TIWGGSGNDD47 pKa = 3.84TIRR50 pKa = 11.84SSGAGVYY57 pKa = 10.51DD58 pKa = 3.98GGTGNDD64 pKa = 3.3YY65 pKa = 11.21VYY67 pKa = 11.06AGLGTQEE74 pKa = 4.28TLRR77 pKa = 11.84GGAGTDD83 pKa = 3.35WLNTTHH89 pKa = 7.24WNGNYY94 pKa = 8.96TIYY97 pKa = 8.03MTTGATNHH105 pKa = 6.17TGEE108 pKa = 5.1SFTQFEE114 pKa = 4.46NLVTGNGNDD123 pKa = 3.96NITGTSGANRR133 pKa = 11.84IYY135 pKa = 10.37TYY137 pKa = 10.99GGNDD141 pKa = 3.13RR142 pKa = 11.84VNAGAGNDD150 pKa = 3.95YY151 pKa = 11.08VWTGDD156 pKa = 3.66GNDD159 pKa = 4.49SIDD162 pKa = 3.6AGSGRR167 pKa = 11.84DD168 pKa = 3.33TVYY171 pKa = 11.04GGNGNDD177 pKa = 4.23TIRR180 pKa = 11.84SSGSGIFDD188 pKa = 3.41GQGGNDD194 pKa = 3.86YY195 pKa = 10.43IYY197 pKa = 11.02AGLGTQEE204 pKa = 4.28TLRR207 pKa = 11.84GGAGTDD213 pKa = 3.35WLNTTHH219 pKa = 7.03WNSDD223 pKa = 3.17YY224 pKa = 10.31TINMVTGATNYY235 pKa = 9.07TGEE238 pKa = 4.48SFTQFEE244 pKa = 4.46NLVTGNGDD252 pKa = 3.6DD253 pKa = 4.72NIIGTADD260 pKa = 3.4GNRR263 pKa = 11.84IYY265 pKa = 11.06TNGGNDD271 pKa = 3.5KK272 pKa = 10.64VDD274 pKa = 3.46AGAGNDD280 pKa = 3.98YY281 pKa = 11.15VWTANGNDD289 pKa = 4.17SIDD292 pKa = 3.63AGSGRR297 pKa = 11.84DD298 pKa = 3.33TVYY301 pKa = 11.04GGNGNDD307 pKa = 4.23TIRR310 pKa = 11.84SSGSGIFDD318 pKa = 3.41GQGDD322 pKa = 4.01NDD324 pKa = 3.74YY325 pKa = 11.39VYY327 pKa = 10.98AGLGAQEE334 pKa = 4.28TLRR337 pKa = 11.84GGAGTDD343 pKa = 3.35WLNTTHH349 pKa = 7.24WNGNYY354 pKa = 8.55TINMTTGATNYY365 pKa = 8.91TGEE368 pKa = 4.48SFTQFEE374 pKa = 4.46NLVTGNGNDD383 pKa = 3.96NITGTSGANRR393 pKa = 11.84IYY395 pKa = 10.7TRR397 pKa = 11.84GGNDD401 pKa = 2.85TVAAGAGNDD410 pKa = 3.95YY411 pKa = 11.18VSGGAGNDD419 pKa = 3.66SLDD422 pKa = 4.05GGSGTNTVYY431 pKa = 10.73GGSGDD436 pKa = 3.86DD437 pKa = 4.04VIRR440 pKa = 11.84SSGRR444 pKa = 11.84GVYY447 pKa = 10.13DD448 pKa = 3.67GQDD451 pKa = 3.66GNDD454 pKa = 4.41LIYY457 pKa = 10.86AGNGTPEE464 pKa = 4.19TLRR467 pKa = 11.84GGSGIDD473 pKa = 3.46TLNTTSFGGDD483 pKa = 3.48YY484 pKa = 10.48NIDD487 pKa = 3.55MATGTTNFSGEE498 pKa = 4.15SFTQFEE504 pKa = 4.47HH505 pKa = 7.03LIAGAGDD512 pKa = 3.48DD513 pKa = 3.88TLAGNSASNRR523 pKa = 11.84ISGGDD528 pKa = 3.35GNDD531 pKa = 3.93RR532 pKa = 11.84ITGLDD537 pKa = 3.78GNDD540 pKa = 3.33TLYY543 pKa = 11.43GDD545 pKa = 4.3NGNDD549 pKa = 3.46YY550 pKa = 11.28LSGGNGNDD558 pKa = 3.95LLFGGNGNDD567 pKa = 3.34TMLGGSGNDD576 pKa = 2.62RR577 pKa = 11.84MYY579 pKa = 11.4GNNGVDD585 pKa = 3.14RR586 pKa = 11.84MFGGSGNDD594 pKa = 3.38YY595 pKa = 10.44MRR597 pKa = 11.84GGNDD601 pKa = 3.02NDD603 pKa = 3.71RR604 pKa = 11.84LYY606 pKa = 11.01GQNGRR611 pKa = 11.84DD612 pKa = 3.53NMYY615 pKa = 11.16GDD617 pKa = 4.62AGNDD621 pKa = 3.05TMYY624 pKa = 11.05GGNQNDD630 pKa = 3.71RR631 pKa = 11.84MDD633 pKa = 4.24GGSGNDD639 pKa = 3.27TMFGQSGDD647 pKa = 3.56DD648 pKa = 3.69RR649 pKa = 11.84ISGRR653 pKa = 11.84LGADD657 pKa = 3.28VMSGGGGEE665 pKa = 4.23DD666 pKa = 3.1TFVFYY671 pKa = 10.41STADD675 pKa = 3.52SPFGNSANYY684 pKa = 10.28DD685 pKa = 4.04RR686 pKa = 11.84ITDD689 pKa = 3.88FQGAGINLLSTIEE702 pKa = 4.19DD703 pKa = 4.31KK704 pKa = 10.93IDD706 pKa = 3.98LSAIDD711 pKa = 4.73ANTGIAGNQAFTFSGTSNGGAGSIWMQNVGSEE743 pKa = 3.52TWLRR747 pKa = 11.84VNTDD751 pKa = 3.06ADD753 pKa = 3.94STPEE757 pKa = 3.6MTIRR761 pKa = 11.84ISDD764 pKa = 3.85GADD767 pKa = 3.09DD768 pKa = 5.98ANDD771 pKa = 3.13YY772 pKa = 8.84WAGDD776 pKa = 4.15FILL779 pKa = 5.43
MM1 pKa = 7.33ATLFGNSGNNTIPGTSGADD20 pKa = 3.78LIYY23 pKa = 10.85GFGGNDD29 pKa = 3.16SLGGNGGRR37 pKa = 11.84DD38 pKa = 4.01TIWGGSGNDD47 pKa = 3.84TIRR50 pKa = 11.84SSGAGVYY57 pKa = 10.51DD58 pKa = 3.98GGTGNDD64 pKa = 3.3YY65 pKa = 11.21VYY67 pKa = 11.06AGLGTQEE74 pKa = 4.28TLRR77 pKa = 11.84GGAGTDD83 pKa = 3.35WLNTTHH89 pKa = 7.24WNGNYY94 pKa = 8.96TIYY97 pKa = 8.03MTTGATNHH105 pKa = 6.17TGEE108 pKa = 5.1SFTQFEE114 pKa = 4.46NLVTGNGNDD123 pKa = 3.96NITGTSGANRR133 pKa = 11.84IYY135 pKa = 10.37TYY137 pKa = 10.99GGNDD141 pKa = 3.13RR142 pKa = 11.84VNAGAGNDD150 pKa = 3.95YY151 pKa = 11.08VWTGDD156 pKa = 3.66GNDD159 pKa = 4.49SIDD162 pKa = 3.6AGSGRR167 pKa = 11.84DD168 pKa = 3.33TVYY171 pKa = 11.04GGNGNDD177 pKa = 4.23TIRR180 pKa = 11.84SSGSGIFDD188 pKa = 3.41GQGGNDD194 pKa = 3.86YY195 pKa = 10.43IYY197 pKa = 11.02AGLGTQEE204 pKa = 4.28TLRR207 pKa = 11.84GGAGTDD213 pKa = 3.35WLNTTHH219 pKa = 7.03WNSDD223 pKa = 3.17YY224 pKa = 10.31TINMVTGATNYY235 pKa = 9.07TGEE238 pKa = 4.48SFTQFEE244 pKa = 4.46NLVTGNGDD252 pKa = 3.6DD253 pKa = 4.72NIIGTADD260 pKa = 3.4GNRR263 pKa = 11.84IYY265 pKa = 11.06TNGGNDD271 pKa = 3.5KK272 pKa = 10.64VDD274 pKa = 3.46AGAGNDD280 pKa = 3.98YY281 pKa = 11.15VWTANGNDD289 pKa = 4.17SIDD292 pKa = 3.63AGSGRR297 pKa = 11.84DD298 pKa = 3.33TVYY301 pKa = 11.04GGNGNDD307 pKa = 4.23TIRR310 pKa = 11.84SSGSGIFDD318 pKa = 3.41GQGDD322 pKa = 4.01NDD324 pKa = 3.74YY325 pKa = 11.39VYY327 pKa = 10.98AGLGAQEE334 pKa = 4.28TLRR337 pKa = 11.84GGAGTDD343 pKa = 3.35WLNTTHH349 pKa = 7.24WNGNYY354 pKa = 8.55TINMTTGATNYY365 pKa = 8.91TGEE368 pKa = 4.48SFTQFEE374 pKa = 4.46NLVTGNGNDD383 pKa = 3.96NITGTSGANRR393 pKa = 11.84IYY395 pKa = 10.7TRR397 pKa = 11.84GGNDD401 pKa = 2.85TVAAGAGNDD410 pKa = 3.95YY411 pKa = 11.18VSGGAGNDD419 pKa = 3.66SLDD422 pKa = 4.05GGSGTNTVYY431 pKa = 10.73GGSGDD436 pKa = 3.86DD437 pKa = 4.04VIRR440 pKa = 11.84SSGRR444 pKa = 11.84GVYY447 pKa = 10.13DD448 pKa = 3.67GQDD451 pKa = 3.66GNDD454 pKa = 4.41LIYY457 pKa = 10.86AGNGTPEE464 pKa = 4.19TLRR467 pKa = 11.84GGSGIDD473 pKa = 3.46TLNTTSFGGDD483 pKa = 3.48YY484 pKa = 10.48NIDD487 pKa = 3.55MATGTTNFSGEE498 pKa = 4.15SFTQFEE504 pKa = 4.47HH505 pKa = 7.03LIAGAGDD512 pKa = 3.48DD513 pKa = 3.88TLAGNSASNRR523 pKa = 11.84ISGGDD528 pKa = 3.35GNDD531 pKa = 3.93RR532 pKa = 11.84ITGLDD537 pKa = 3.78GNDD540 pKa = 3.33TLYY543 pKa = 11.43GDD545 pKa = 4.3NGNDD549 pKa = 3.46YY550 pKa = 11.28LSGGNGNDD558 pKa = 3.95LLFGGNGNDD567 pKa = 3.34TMLGGSGNDD576 pKa = 2.62RR577 pKa = 11.84MYY579 pKa = 11.4GNNGVDD585 pKa = 3.14RR586 pKa = 11.84MFGGSGNDD594 pKa = 3.38YY595 pKa = 10.44MRR597 pKa = 11.84GGNDD601 pKa = 3.02NDD603 pKa = 3.71RR604 pKa = 11.84LYY606 pKa = 11.01GQNGRR611 pKa = 11.84DD612 pKa = 3.53NMYY615 pKa = 11.16GDD617 pKa = 4.62AGNDD621 pKa = 3.05TMYY624 pKa = 11.05GGNQNDD630 pKa = 3.71RR631 pKa = 11.84MDD633 pKa = 4.24GGSGNDD639 pKa = 3.27TMFGQSGDD647 pKa = 3.56DD648 pKa = 3.69RR649 pKa = 11.84ISGRR653 pKa = 11.84LGADD657 pKa = 3.28VMSGGGGEE665 pKa = 4.23DD666 pKa = 3.1TFVFYY671 pKa = 10.41STADD675 pKa = 3.52SPFGNSANYY684 pKa = 10.28DD685 pKa = 4.04RR686 pKa = 11.84ITDD689 pKa = 3.88FQGAGINLLSTIEE702 pKa = 4.19DD703 pKa = 4.31KK704 pKa = 10.93IDD706 pKa = 3.98LSAIDD711 pKa = 4.73ANTGIAGNQAFTFSGTSNGGAGSIWMQNVGSEE743 pKa = 3.52TWLRR747 pKa = 11.84VNTDD751 pKa = 3.06ADD753 pKa = 3.94STPEE757 pKa = 3.6MTIRR761 pKa = 11.84ISDD764 pKa = 3.85GADD767 pKa = 3.09DD768 pKa = 5.98ANDD771 pKa = 3.13YY772 pKa = 8.84WAGDD776 pKa = 4.15FILL779 pKa = 5.43
Molecular weight: 80.18 kDa
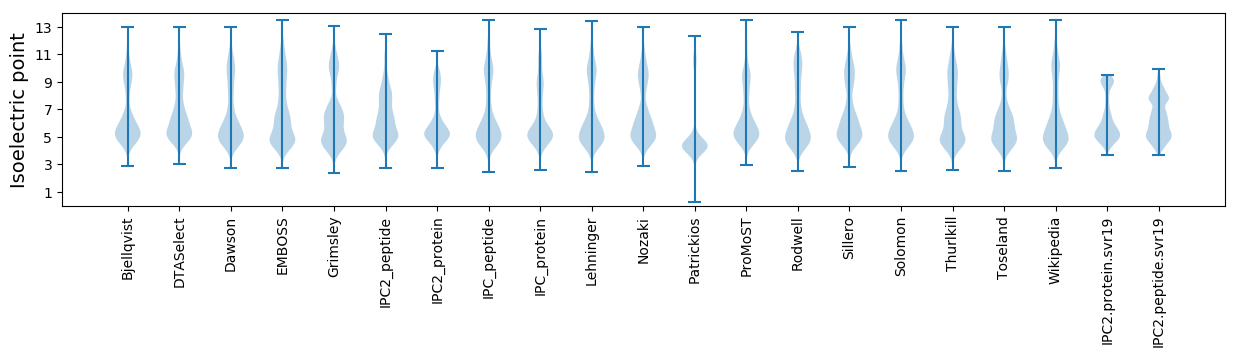
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9Y8F2|A0A1H9Y8F2_9RHOB Protein RodZ contains Xre-like HTH and DUF4115 domains OS=Paracoccus homiensis OX=364199 GN=SAMN04489858_10158 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1177105 |
27 |
4492 |
307.5 |
33.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.61 ± 0.058 | 0.867 ± 0.013 |
6.428 ± 0.041 | 5.237 ± 0.043 |
3.548 ± 0.029 | 8.939 ± 0.083 |
2.069 ± 0.023 | 5.288 ± 0.03 |
2.734 ± 0.033 | 10.078 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.852 ± 0.023 | 2.466 ± 0.024 |
5.222 ± 0.03 | 3.654 ± 0.026 |
7.184 ± 0.047 | 5.027 ± 0.034 |
5.265 ± 0.026 | 7.0 ± 0.035 |
1.459 ± 0.019 | 2.074 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
