
Kocuria flava
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Kocuria
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
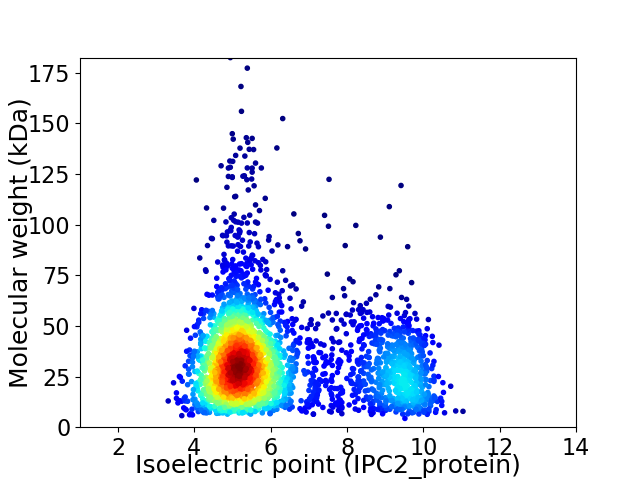
Virtual 2D-PAGE plot for 3024 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U3GQ82|A0A0U3GQ82_9MICC Alpha/beta hydrolase OS=Kocuria flava OX=446860 GN=AS188_15755 PE=4 SV=1
MM1 pKa = 7.41AASTVPLLVLTGCASDD17 pKa = 4.24AGAEE21 pKa = 3.92AAATGVVDD29 pKa = 3.1YY30 pKa = 10.59WMWDD34 pKa = 3.58TNQLPAYY41 pKa = 7.67QKK43 pKa = 9.43CAEE46 pKa = 4.27MFEE49 pKa = 5.04AEE51 pKa = 5.52HH52 pKa = 7.29PDD54 pKa = 2.91QDD56 pKa = 4.58VRR58 pKa = 11.84ITQIGWGDD66 pKa = 3.62YY67 pKa = 6.46WTKK70 pKa = 9.96LTAGFIAGTGPDD82 pKa = 3.23VFTNHH87 pKa = 5.29VSKK90 pKa = 11.05YY91 pKa = 8.13PQFAEE96 pKa = 4.34LEE98 pKa = 4.25VLAPLEE104 pKa = 4.14EE105 pKa = 4.82HH106 pKa = 7.02PATAALEE113 pKa = 4.24PADD116 pKa = 4.04YY117 pKa = 10.88QDD119 pKa = 4.22GLVDD123 pKa = 4.49LWTGPDD129 pKa = 3.02GHH131 pKa = 6.62QYY133 pKa = 10.72GVPKK137 pKa = 10.6DD138 pKa = 3.36WDD140 pKa = 4.05TVAVLYY146 pKa = 10.92NEE148 pKa = 4.23EE149 pKa = 4.01LVAEE153 pKa = 4.85AGLSPEE159 pKa = 4.99DD160 pKa = 3.77LEE162 pKa = 4.42GWRR165 pKa = 11.84WNPEE169 pKa = 3.9DD170 pKa = 3.93GGSFEE175 pKa = 5.97DD176 pKa = 5.0LVAHH180 pKa = 6.34LTIDD184 pKa = 3.69EE185 pKa = 4.33NGVRR189 pKa = 11.84GDD191 pKa = 3.62EE192 pKa = 4.06PGFDD196 pKa = 3.72KK197 pKa = 11.38DD198 pKa = 3.42HH199 pKa = 6.0VAVYY203 pKa = 10.8GLGISEE209 pKa = 5.02AGGSTAGQGQWSAFAASNGWRR230 pKa = 11.84VTDD233 pKa = 3.6EE234 pKa = 4.45PLWGDD239 pKa = 4.26HH240 pKa = 6.3YY241 pKa = 11.76NVDD244 pKa = 4.53DD245 pKa = 4.52PALHH249 pKa = 6.4EE250 pKa = 4.16TLDD253 pKa = 3.61WYY255 pKa = 11.1FGLTDD260 pKa = 3.62KK261 pKa = 11.37GYY263 pKa = 9.0MPAYY267 pKa = 9.47GQFNAAEE274 pKa = 4.31GVNAQLASGSAAMVLDD290 pKa = 4.43GAWMISTYY298 pKa = 11.38AAMDD302 pKa = 5.1DD303 pKa = 3.71ISVGTARR310 pKa = 11.84TPVGPDD316 pKa = 3.18GTSTSMMNGLADD328 pKa = 4.88SIVKK332 pKa = 8.91DD333 pKa = 3.46TDD335 pKa = 3.52NPEE338 pKa = 4.58GAAQWAAFLGSAQCQQEE355 pKa = 4.27VARR358 pKa = 11.84AGVVFPARR366 pKa = 11.84QDD368 pKa = 3.43ATPIAADD375 pKa = 4.58AYY377 pKa = 9.16RR378 pKa = 11.84QMGIDD383 pKa = 4.12PEE385 pKa = 4.27PFIAPVEE392 pKa = 4.26EE393 pKa = 4.74GSTVYY398 pKa = 10.49FPVTKK403 pKa = 10.67FGADD407 pKa = 3.19VSALMTPALEE417 pKa = 5.13DD418 pKa = 3.0IYY420 pKa = 11.36ANRR423 pKa = 11.84VPASTLTGINDD434 pKa = 5.16AINVLFEE441 pKa = 4.03TDD443 pKa = 3.28EE444 pKa = 4.64TEE446 pKa = 4.23GSS448 pKa = 3.46
MM1 pKa = 7.41AASTVPLLVLTGCASDD17 pKa = 4.24AGAEE21 pKa = 3.92AAATGVVDD29 pKa = 3.1YY30 pKa = 10.59WMWDD34 pKa = 3.58TNQLPAYY41 pKa = 7.67QKK43 pKa = 9.43CAEE46 pKa = 4.27MFEE49 pKa = 5.04AEE51 pKa = 5.52HH52 pKa = 7.29PDD54 pKa = 2.91QDD56 pKa = 4.58VRR58 pKa = 11.84ITQIGWGDD66 pKa = 3.62YY67 pKa = 6.46WTKK70 pKa = 9.96LTAGFIAGTGPDD82 pKa = 3.23VFTNHH87 pKa = 5.29VSKK90 pKa = 11.05YY91 pKa = 8.13PQFAEE96 pKa = 4.34LEE98 pKa = 4.25VLAPLEE104 pKa = 4.14EE105 pKa = 4.82HH106 pKa = 7.02PATAALEE113 pKa = 4.24PADD116 pKa = 4.04YY117 pKa = 10.88QDD119 pKa = 4.22GLVDD123 pKa = 4.49LWTGPDD129 pKa = 3.02GHH131 pKa = 6.62QYY133 pKa = 10.72GVPKK137 pKa = 10.6DD138 pKa = 3.36WDD140 pKa = 4.05TVAVLYY146 pKa = 10.92NEE148 pKa = 4.23EE149 pKa = 4.01LVAEE153 pKa = 4.85AGLSPEE159 pKa = 4.99DD160 pKa = 3.77LEE162 pKa = 4.42GWRR165 pKa = 11.84WNPEE169 pKa = 3.9DD170 pKa = 3.93GGSFEE175 pKa = 5.97DD176 pKa = 5.0LVAHH180 pKa = 6.34LTIDD184 pKa = 3.69EE185 pKa = 4.33NGVRR189 pKa = 11.84GDD191 pKa = 3.62EE192 pKa = 4.06PGFDD196 pKa = 3.72KK197 pKa = 11.38DD198 pKa = 3.42HH199 pKa = 6.0VAVYY203 pKa = 10.8GLGISEE209 pKa = 5.02AGGSTAGQGQWSAFAASNGWRR230 pKa = 11.84VTDD233 pKa = 3.6EE234 pKa = 4.45PLWGDD239 pKa = 4.26HH240 pKa = 6.3YY241 pKa = 11.76NVDD244 pKa = 4.53DD245 pKa = 4.52PALHH249 pKa = 6.4EE250 pKa = 4.16TLDD253 pKa = 3.61WYY255 pKa = 11.1FGLTDD260 pKa = 3.62KK261 pKa = 11.37GYY263 pKa = 9.0MPAYY267 pKa = 9.47GQFNAAEE274 pKa = 4.31GVNAQLASGSAAMVLDD290 pKa = 4.43GAWMISTYY298 pKa = 11.38AAMDD302 pKa = 5.1DD303 pKa = 3.71ISVGTARR310 pKa = 11.84TPVGPDD316 pKa = 3.18GTSTSMMNGLADD328 pKa = 4.88SIVKK332 pKa = 8.91DD333 pKa = 3.46TDD335 pKa = 3.52NPEE338 pKa = 4.58GAAQWAAFLGSAQCQQEE355 pKa = 4.27VARR358 pKa = 11.84AGVVFPARR366 pKa = 11.84QDD368 pKa = 3.43ATPIAADD375 pKa = 4.58AYY377 pKa = 9.16RR378 pKa = 11.84QMGIDD383 pKa = 4.12PEE385 pKa = 4.27PFIAPVEE392 pKa = 4.26EE393 pKa = 4.74GSTVYY398 pKa = 10.49FPVTKK403 pKa = 10.67FGADD407 pKa = 3.19VSALMTPALEE417 pKa = 5.13DD418 pKa = 3.0IYY420 pKa = 11.36ANRR423 pKa = 11.84VPASTLTGINDD434 pKa = 5.16AINVLFEE441 pKa = 4.03TDD443 pKa = 3.28EE444 pKa = 4.64TEE446 pKa = 4.23GSS448 pKa = 3.46
Molecular weight: 47.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U3HVS4|A0A0U3HVS4_9MICC Uncharacterized protein OS=Kocuria flava OX=446860 GN=AS188_03815 PE=4 SV=1
MM1 pKa = 7.08VPPWSSTIVRR11 pKa = 11.84ATARR15 pKa = 11.84PSPRR19 pKa = 11.84PGIALIRR26 pKa = 11.84ALRR29 pKa = 11.84LRR31 pKa = 11.84PKK33 pKa = 10.43RR34 pKa = 11.84SRR36 pKa = 11.84TRR38 pKa = 11.84SASSAGRR45 pKa = 11.84PIPRR49 pKa = 11.84SATRR53 pKa = 11.84SRR55 pKa = 11.84AVSPSRR61 pKa = 11.84STRR64 pKa = 11.84TVTGPPSGEE73 pKa = 3.79
MM1 pKa = 7.08VPPWSSTIVRR11 pKa = 11.84ATARR15 pKa = 11.84PSPRR19 pKa = 11.84PGIALIRR26 pKa = 11.84ALRR29 pKa = 11.84LRR31 pKa = 11.84PKK33 pKa = 10.43RR34 pKa = 11.84SRR36 pKa = 11.84TRR38 pKa = 11.84SASSAGRR45 pKa = 11.84PIPRR49 pKa = 11.84SATRR53 pKa = 11.84SRR55 pKa = 11.84AVSPSRR61 pKa = 11.84STRR64 pKa = 11.84TVTGPPSGEE73 pKa = 3.79
Molecular weight: 7.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
990024 |
37 |
1667 |
327.4 |
34.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.19 ± 0.079 | 0.64 ± 0.012 |
5.698 ± 0.038 | 6.304 ± 0.047 |
2.722 ± 0.025 | 9.564 ± 0.046 |
2.355 ± 0.023 | 2.975 ± 0.037 |
1.616 ± 0.027 | 10.536 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.706 ± 0.017 | 1.612 ± 0.023 |
6.211 ± 0.044 | 2.753 ± 0.024 |
8.176 ± 0.058 | 4.515 ± 0.03 |
5.807 ± 0.032 | 9.262 ± 0.043 |
1.504 ± 0.019 | 1.854 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
