
Agromyces sp. MMS17-SY077
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces; unclassified Agromyces
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
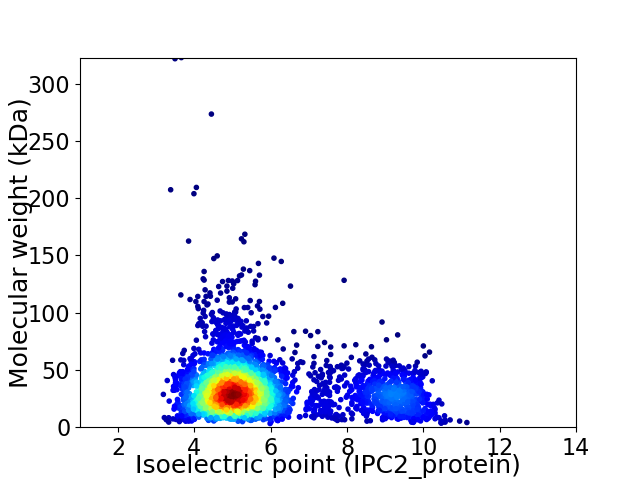
Virtual 2D-PAGE plot for 3503 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I4NZX8|A0A6I4NZX8_9MICO Uncharacterized protein OS=Agromyces sp. MMS17-SY077 OX=2662446 GN=GB864_09465 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.5SKK4 pKa = 10.76LLATAGVVALSAAALTGCSTASGGDD29 pKa = 3.57SAKK32 pKa = 10.65ADD34 pKa = 3.71CTNEE38 pKa = 3.71IVNPDD43 pKa = 3.3ATQVSVWAWYY53 pKa = 8.14PAFEE57 pKa = 4.24EE58 pKa = 4.65VVDD61 pKa = 5.01LFNEE65 pKa = 4.14SHH67 pKa = 7.85DD68 pKa = 4.13DD69 pKa = 4.6VQICWTNAGQGNDD82 pKa = 3.61EE83 pKa = 4.1YY84 pKa = 10.83TKK86 pKa = 10.76FSTAIEE92 pKa = 4.47AGSGAPDD99 pKa = 3.59VIMLEE104 pKa = 4.27SEE106 pKa = 4.38VLSSFSIRR114 pKa = 11.84KK115 pKa = 9.16ALVDD119 pKa = 3.6LTEE122 pKa = 4.18YY123 pKa = 10.9GAADD127 pKa = 3.58VQDD130 pKa = 5.45DD131 pKa = 4.21YY132 pKa = 12.0TAGAWKK138 pKa = 10.3DD139 pKa = 3.79VSSGDD144 pKa = 3.36AVYY147 pKa = 10.34AIPVDD152 pKa = 4.16GGPMGMLYY160 pKa = 10.4RR161 pKa = 11.84KK162 pKa = 10.15DD163 pKa = 4.04ILDD166 pKa = 3.37QYY168 pKa = 11.01GIAAPTTWDD177 pKa = 3.37EE178 pKa = 4.43FKK180 pKa = 11.05AAAEE184 pKa = 3.95ALKK187 pKa = 10.7AAGAPGVLADD197 pKa = 4.64FPTNGRR203 pKa = 11.84AYY205 pKa = 10.15NQALFAQAGSVPFVYY220 pKa = 10.67DD221 pKa = 3.23NANPTEE227 pKa = 3.96IGIDD231 pKa = 3.73VNDD234 pKa = 4.04EE235 pKa = 3.73GSKK238 pKa = 10.39QVLSYY243 pKa = 10.46WNEE246 pKa = 3.67LVQDD250 pKa = 4.14GLVATDD256 pKa = 4.18DD257 pKa = 4.86AFTADD262 pKa = 4.2YY263 pKa = 7.76NTKK266 pKa = 10.53LVDD269 pKa = 3.71GSYY272 pKa = 10.7AIYY275 pKa = 10.3VAAAWGPGYY284 pKa = 10.88LSGLADD290 pKa = 4.16ADD292 pKa = 4.77SDD294 pKa = 4.57AEE296 pKa = 4.23WVAAPVPQWDD306 pKa = 3.61AANPVQINWGGSTFAVTSQAKK327 pKa = 10.07DD328 pKa = 3.06KK329 pKa = 10.68EE330 pKa = 4.28AAATVAKK337 pKa = 10.04EE338 pKa = 3.98VFGTEE343 pKa = 4.19EE344 pKa = 3.41AWKK347 pKa = 10.41IGIEE351 pKa = 3.97QGALFPLWKK360 pKa = 9.9PILEE364 pKa = 4.04SDD366 pKa = 3.72YY367 pKa = 10.99FRR369 pKa = 11.84DD370 pKa = 3.23LEE372 pKa = 4.32YY373 pKa = 10.45PFFGGQQINKK383 pKa = 9.86DD384 pKa = 3.63VFLAAAAGYY393 pKa = 10.22EE394 pKa = 4.24GFTFSPFQNYY404 pKa = 10.17AYY406 pKa = 10.31DD407 pKa = 3.62QLTEE411 pKa = 3.67QLYY414 pKa = 11.37AMVQGEE420 pKa = 4.35KK421 pKa = 10.66SSDD424 pKa = 3.38QALDD428 pKa = 3.79DD429 pKa = 4.31LQASLEE435 pKa = 4.2QYY437 pKa = 10.69AGEE440 pKa = 4.72QGFTLQQ446 pKa = 4.51
MM1 pKa = 7.54KK2 pKa = 10.5SKK4 pKa = 10.76LLATAGVVALSAAALTGCSTASGGDD29 pKa = 3.57SAKK32 pKa = 10.65ADD34 pKa = 3.71CTNEE38 pKa = 3.71IVNPDD43 pKa = 3.3ATQVSVWAWYY53 pKa = 8.14PAFEE57 pKa = 4.24EE58 pKa = 4.65VVDD61 pKa = 5.01LFNEE65 pKa = 4.14SHH67 pKa = 7.85DD68 pKa = 4.13DD69 pKa = 4.6VQICWTNAGQGNDD82 pKa = 3.61EE83 pKa = 4.1YY84 pKa = 10.83TKK86 pKa = 10.76FSTAIEE92 pKa = 4.47AGSGAPDD99 pKa = 3.59VIMLEE104 pKa = 4.27SEE106 pKa = 4.38VLSSFSIRR114 pKa = 11.84KK115 pKa = 9.16ALVDD119 pKa = 3.6LTEE122 pKa = 4.18YY123 pKa = 10.9GAADD127 pKa = 3.58VQDD130 pKa = 5.45DD131 pKa = 4.21YY132 pKa = 12.0TAGAWKK138 pKa = 10.3DD139 pKa = 3.79VSSGDD144 pKa = 3.36AVYY147 pKa = 10.34AIPVDD152 pKa = 4.16GGPMGMLYY160 pKa = 10.4RR161 pKa = 11.84KK162 pKa = 10.15DD163 pKa = 4.04ILDD166 pKa = 3.37QYY168 pKa = 11.01GIAAPTTWDD177 pKa = 3.37EE178 pKa = 4.43FKK180 pKa = 11.05AAAEE184 pKa = 3.95ALKK187 pKa = 10.7AAGAPGVLADD197 pKa = 4.64FPTNGRR203 pKa = 11.84AYY205 pKa = 10.15NQALFAQAGSVPFVYY220 pKa = 10.67DD221 pKa = 3.23NANPTEE227 pKa = 3.96IGIDD231 pKa = 3.73VNDD234 pKa = 4.04EE235 pKa = 3.73GSKK238 pKa = 10.39QVLSYY243 pKa = 10.46WNEE246 pKa = 3.67LVQDD250 pKa = 4.14GLVATDD256 pKa = 4.18DD257 pKa = 4.86AFTADD262 pKa = 4.2YY263 pKa = 7.76NTKK266 pKa = 10.53LVDD269 pKa = 3.71GSYY272 pKa = 10.7AIYY275 pKa = 10.3VAAAWGPGYY284 pKa = 10.88LSGLADD290 pKa = 4.16ADD292 pKa = 4.77SDD294 pKa = 4.57AEE296 pKa = 4.23WVAAPVPQWDD306 pKa = 3.61AANPVQINWGGSTFAVTSQAKK327 pKa = 10.07DD328 pKa = 3.06KK329 pKa = 10.68EE330 pKa = 4.28AAATVAKK337 pKa = 10.04EE338 pKa = 3.98VFGTEE343 pKa = 4.19EE344 pKa = 3.41AWKK347 pKa = 10.41IGIEE351 pKa = 3.97QGALFPLWKK360 pKa = 9.9PILEE364 pKa = 4.04SDD366 pKa = 3.72YY367 pKa = 10.99FRR369 pKa = 11.84DD370 pKa = 3.23LEE372 pKa = 4.32YY373 pKa = 10.45PFFGGQQINKK383 pKa = 9.86DD384 pKa = 3.63VFLAAAAGYY393 pKa = 10.22EE394 pKa = 4.24GFTFSPFQNYY404 pKa = 10.17AYY406 pKa = 10.31DD407 pKa = 3.62QLTEE411 pKa = 3.67QLYY414 pKa = 11.37AMVQGEE420 pKa = 4.35KK421 pKa = 10.66SSDD424 pKa = 3.38QALDD428 pKa = 3.79DD429 pKa = 4.31LQASLEE435 pKa = 4.2QYY437 pKa = 10.69AGEE440 pKa = 4.72QGFTLQQ446 pKa = 4.51
Molecular weight: 47.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I4P455|A0A6I4P455_9MICO Flagellar protein FlgN OS=Agromyces sp. MMS17-SY077 OX=2662446 GN=GB864_06500 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1161124 |
29 |
3243 |
331.5 |
35.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.62 ± 0.066 | 0.444 ± 0.009 |
6.31 ± 0.042 | 5.866 ± 0.044 |
3.187 ± 0.024 | 9.449 ± 0.032 |
1.837 ± 0.022 | 4.212 ± 0.031 |
1.699 ± 0.029 | 10.302 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.512 ± 0.016 | 1.773 ± 0.024 |
5.627 ± 0.033 | 2.43 ± 0.025 |
7.369 ± 0.053 | 5.21 ± 0.035 |
5.625 ± 0.046 | 9.058 ± 0.039 |
1.543 ± 0.02 | 1.924 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
