
Gracilibacillus kekensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Gracilibacillus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
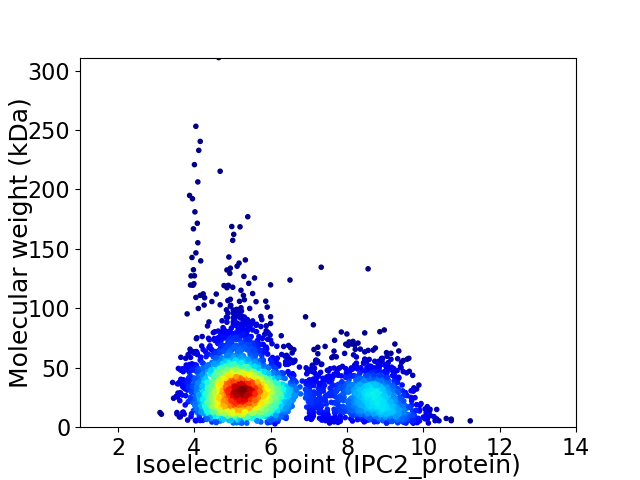
Virtual 2D-PAGE plot for 3690 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
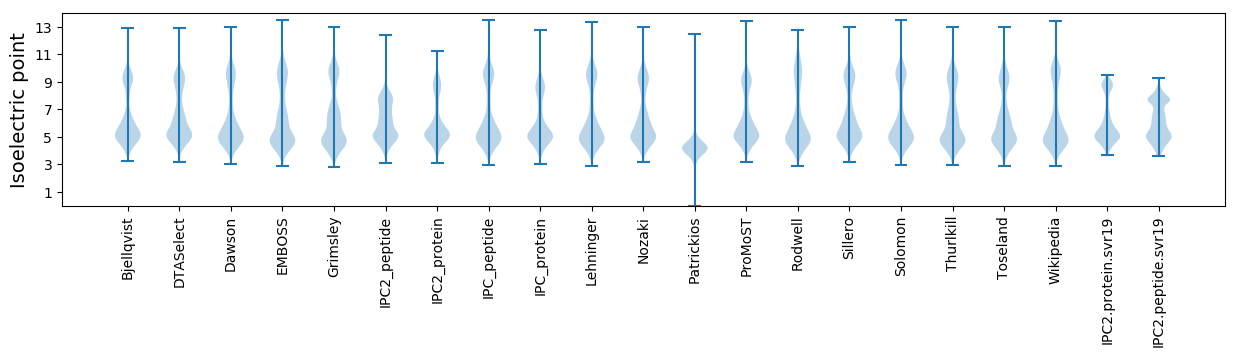
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M7QXB6|A0A1M7QXB6_9BACI Uncharacterized protein OS=Gracilibacillus kekensis OX=1027249 GN=SAMN05216179_3717 PE=4 SV=1
MM1 pKa = 7.55EE2 pKa = 5.46KK3 pKa = 10.46SKK5 pKa = 11.34SLLLLLFTLLFSVMLAACSDD25 pKa = 4.17DD26 pKa = 3.96SDD28 pKa = 4.47SSSSSEE34 pKa = 3.94GNGNDD39 pKa = 3.25SSSEE43 pKa = 4.14TEE45 pKa = 3.79SDD47 pKa = 3.2GGEE50 pKa = 3.68ASGDD54 pKa = 3.58SEE56 pKa = 4.51EE57 pKa = 4.8PYY59 pKa = 10.36EE60 pKa = 4.49ISIMTNAYY68 pKa = 8.46TPEE71 pKa = 4.75PPTPEE76 pKa = 3.91SPAWQAIEE84 pKa = 5.39DD85 pKa = 4.02FTNTEE90 pKa = 3.59LDD92 pKa = 3.07ITYY95 pKa = 9.87VPSSNYY101 pKa = 9.78DD102 pKa = 3.04EE103 pKa = 5.06RR104 pKa = 11.84FNITLASGDD113 pKa = 4.4LPSMILTNKK122 pKa = 8.89TSSFINAVNDD132 pKa = 3.85GAFWDD137 pKa = 3.99LTDD140 pKa = 4.7YY141 pKa = 11.58LDD143 pKa = 5.26DD144 pKa = 4.31YY145 pKa = 11.84EE146 pKa = 5.77NLSQMNDD153 pKa = 2.6IVKK156 pKa = 10.69NNISIDD162 pKa = 3.36GKK164 pKa = 10.66IYY166 pKa = 10.01GLYY169 pKa = 9.73RR170 pKa = 11.84ARR172 pKa = 11.84PLGRR176 pKa = 11.84MAVSIRR182 pKa = 11.84QDD184 pKa = 2.78WLDD187 pKa = 3.61NLGMEE192 pKa = 4.62TPTTTEE198 pKa = 3.18EE199 pKa = 4.8FYY201 pKa = 11.36DD202 pKa = 3.44VMYY205 pKa = 10.95AFTHH209 pKa = 7.29DD210 pKa = 4.67DD211 pKa = 3.64PDD213 pKa = 5.51GNGQDD218 pKa = 3.95DD219 pKa = 4.16TLGTIVSEE227 pKa = 4.82YY228 pKa = 9.57PGPWDD233 pKa = 5.1IMQTWFGVPNKK244 pKa = 9.5WGEE247 pKa = 4.19NEE249 pKa = 5.08DD250 pKa = 3.68GTLYY254 pKa = 10.32PYY256 pKa = 10.6FQDD259 pKa = 3.62PAYY262 pKa = 9.98KK263 pKa = 10.21EE264 pKa = 3.73ALDD267 pKa = 3.89YY268 pKa = 10.37FKK270 pKa = 11.14RR271 pKa = 11.84MYY273 pKa = 11.05DD274 pKa = 3.54DD275 pKa = 5.29GLVNDD280 pKa = 5.44DD281 pKa = 4.32FAVMDD286 pKa = 4.06PAKK289 pKa = 10.27WHH291 pKa = 6.41DD292 pKa = 3.46AYY294 pKa = 11.92VNGEE298 pKa = 4.03AGTVVDD304 pKa = 4.3VADD307 pKa = 3.74AASRR311 pKa = 11.84NRR313 pKa = 11.84DD314 pKa = 3.15KK315 pKa = 10.79MVEE318 pKa = 3.98ADD320 pKa = 3.69PSLEE324 pKa = 3.8GSVDD328 pKa = 3.21VFGAVEE334 pKa = 4.43GPNGLHH340 pKa = 6.63HH341 pKa = 7.2LPTSGYY347 pKa = 10.09NMMYY351 pKa = 10.24AISTTAVEE359 pKa = 4.58TEE361 pKa = 4.19EE362 pKa = 4.42EE363 pKa = 4.12LHH365 pKa = 7.04KK366 pKa = 10.91VLQFMDD372 pKa = 3.86DD373 pKa = 3.43MNTEE377 pKa = 4.25EE378 pKa = 4.34GQKK381 pKa = 10.41LGYY384 pKa = 10.24NGVEE388 pKa = 4.07GTHH391 pKa = 6.96YY392 pKa = 10.72EE393 pKa = 4.16MKK395 pKa = 10.51DD396 pKa = 3.36GEE398 pKa = 4.62FVPTTDD404 pKa = 2.7QALIYY409 pKa = 10.0EE410 pKa = 4.4YY411 pKa = 11.03EE412 pKa = 4.49DD413 pKa = 3.93LNQLLMFVPEE423 pKa = 4.38DD424 pKa = 3.71RR425 pKa = 11.84YY426 pKa = 9.74LTEE429 pKa = 4.73PVDD432 pKa = 4.58DD433 pKa = 3.88LTKK436 pKa = 10.58KK437 pKa = 10.31EE438 pKa = 4.11EE439 pKa = 3.95EE440 pKa = 4.02VLAEE444 pKa = 3.98NEE446 pKa = 4.32EE447 pKa = 4.38IVVGNPAEE455 pKa = 4.13PLVSEE460 pKa = 4.77VYY462 pKa = 10.28SRR464 pKa = 11.84QGQQLDD470 pKa = 4.21NIILDD475 pKa = 3.4ARR477 pKa = 11.84VQYY480 pKa = 10.53IVGQIDD486 pKa = 3.5EE487 pKa = 5.04AGLEE491 pKa = 4.13EE492 pKa = 6.06AEE494 pKa = 4.19QLWMNSGGKK503 pKa = 9.89EE504 pKa = 3.9YY505 pKa = 10.2IEE507 pKa = 5.52EE508 pKa = 4.04INQLYY513 pKa = 9.78QEE515 pKa = 4.42SQQ517 pKa = 2.89
MM1 pKa = 7.55EE2 pKa = 5.46KK3 pKa = 10.46SKK5 pKa = 11.34SLLLLLFTLLFSVMLAACSDD25 pKa = 4.17DD26 pKa = 3.96SDD28 pKa = 4.47SSSSSEE34 pKa = 3.94GNGNDD39 pKa = 3.25SSSEE43 pKa = 4.14TEE45 pKa = 3.79SDD47 pKa = 3.2GGEE50 pKa = 3.68ASGDD54 pKa = 3.58SEE56 pKa = 4.51EE57 pKa = 4.8PYY59 pKa = 10.36EE60 pKa = 4.49ISIMTNAYY68 pKa = 8.46TPEE71 pKa = 4.75PPTPEE76 pKa = 3.91SPAWQAIEE84 pKa = 5.39DD85 pKa = 4.02FTNTEE90 pKa = 3.59LDD92 pKa = 3.07ITYY95 pKa = 9.87VPSSNYY101 pKa = 9.78DD102 pKa = 3.04EE103 pKa = 5.06RR104 pKa = 11.84FNITLASGDD113 pKa = 4.4LPSMILTNKK122 pKa = 8.89TSSFINAVNDD132 pKa = 3.85GAFWDD137 pKa = 3.99LTDD140 pKa = 4.7YY141 pKa = 11.58LDD143 pKa = 5.26DD144 pKa = 4.31YY145 pKa = 11.84EE146 pKa = 5.77NLSQMNDD153 pKa = 2.6IVKK156 pKa = 10.69NNISIDD162 pKa = 3.36GKK164 pKa = 10.66IYY166 pKa = 10.01GLYY169 pKa = 9.73RR170 pKa = 11.84ARR172 pKa = 11.84PLGRR176 pKa = 11.84MAVSIRR182 pKa = 11.84QDD184 pKa = 2.78WLDD187 pKa = 3.61NLGMEE192 pKa = 4.62TPTTTEE198 pKa = 3.18EE199 pKa = 4.8FYY201 pKa = 11.36DD202 pKa = 3.44VMYY205 pKa = 10.95AFTHH209 pKa = 7.29DD210 pKa = 4.67DD211 pKa = 3.64PDD213 pKa = 5.51GNGQDD218 pKa = 3.95DD219 pKa = 4.16TLGTIVSEE227 pKa = 4.82YY228 pKa = 9.57PGPWDD233 pKa = 5.1IMQTWFGVPNKK244 pKa = 9.5WGEE247 pKa = 4.19NEE249 pKa = 5.08DD250 pKa = 3.68GTLYY254 pKa = 10.32PYY256 pKa = 10.6FQDD259 pKa = 3.62PAYY262 pKa = 9.98KK263 pKa = 10.21EE264 pKa = 3.73ALDD267 pKa = 3.89YY268 pKa = 10.37FKK270 pKa = 11.14RR271 pKa = 11.84MYY273 pKa = 11.05DD274 pKa = 3.54DD275 pKa = 5.29GLVNDD280 pKa = 5.44DD281 pKa = 4.32FAVMDD286 pKa = 4.06PAKK289 pKa = 10.27WHH291 pKa = 6.41DD292 pKa = 3.46AYY294 pKa = 11.92VNGEE298 pKa = 4.03AGTVVDD304 pKa = 4.3VADD307 pKa = 3.74AASRR311 pKa = 11.84NRR313 pKa = 11.84DD314 pKa = 3.15KK315 pKa = 10.79MVEE318 pKa = 3.98ADD320 pKa = 3.69PSLEE324 pKa = 3.8GSVDD328 pKa = 3.21VFGAVEE334 pKa = 4.43GPNGLHH340 pKa = 6.63HH341 pKa = 7.2LPTSGYY347 pKa = 10.09NMMYY351 pKa = 10.24AISTTAVEE359 pKa = 4.58TEE361 pKa = 4.19EE362 pKa = 4.42EE363 pKa = 4.12LHH365 pKa = 7.04KK366 pKa = 10.91VLQFMDD372 pKa = 3.86DD373 pKa = 3.43MNTEE377 pKa = 4.25EE378 pKa = 4.34GQKK381 pKa = 10.41LGYY384 pKa = 10.24NGVEE388 pKa = 4.07GTHH391 pKa = 6.96YY392 pKa = 10.72EE393 pKa = 4.16MKK395 pKa = 10.51DD396 pKa = 3.36GEE398 pKa = 4.62FVPTTDD404 pKa = 2.7QALIYY409 pKa = 10.0EE410 pKa = 4.4YY411 pKa = 11.03EE412 pKa = 4.49DD413 pKa = 3.93LNQLLMFVPEE423 pKa = 4.38DD424 pKa = 3.71RR425 pKa = 11.84YY426 pKa = 9.74LTEE429 pKa = 4.73PVDD432 pKa = 4.58DD433 pKa = 3.88LTKK436 pKa = 10.58KK437 pKa = 10.31EE438 pKa = 4.11EE439 pKa = 3.95EE440 pKa = 4.02VLAEE444 pKa = 3.98NEE446 pKa = 4.32EE447 pKa = 4.38IVVGNPAEE455 pKa = 4.13PLVSEE460 pKa = 4.77VYY462 pKa = 10.28SRR464 pKa = 11.84QGQQLDD470 pKa = 4.21NIILDD475 pKa = 3.4ARR477 pKa = 11.84VQYY480 pKa = 10.53IVGQIDD486 pKa = 3.5EE487 pKa = 5.04AGLEE491 pKa = 4.13EE492 pKa = 6.06AEE494 pKa = 4.19QLWMNSGGKK503 pKa = 9.89EE504 pKa = 3.9YY505 pKa = 10.2IEE507 pKa = 5.52EE508 pKa = 4.04INQLYY513 pKa = 9.78QEE515 pKa = 4.42SQQ517 pKa = 2.89
Molecular weight: 57.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M7QP32|A0A1M7QP32_9BACI Uncharacterized conserved protein YybS DUF2232 family OS=Gracilibacillus kekensis OX=1027249 GN=SAMN05216179_3417 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 8.75VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.2KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 8.75VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.2KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1118330 |
25 |
2738 |
303.1 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.449 ± 0.037 | 0.6 ± 0.011 |
5.591 ± 0.04 | 7.736 ± 0.053 |
4.587 ± 0.034 | 6.345 ± 0.042 |
2.119 ± 0.021 | 8.287 ± 0.042 |
6.667 ± 0.045 | 9.504 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.682 ± 0.021 | 4.883 ± 0.031 |
3.433 ± 0.023 | 4.234 ± 0.033 |
3.672 ± 0.025 | 6.017 ± 0.03 |
5.554 ± 0.027 | 6.784 ± 0.029 |
1.126 ± 0.017 | 3.73 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
