
Sanxia picorna-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
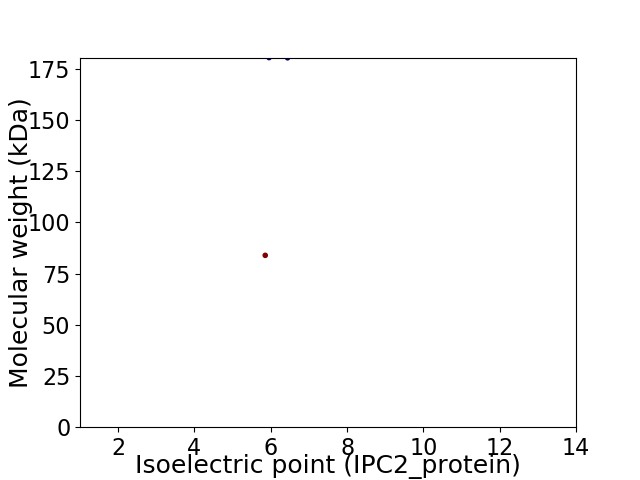
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
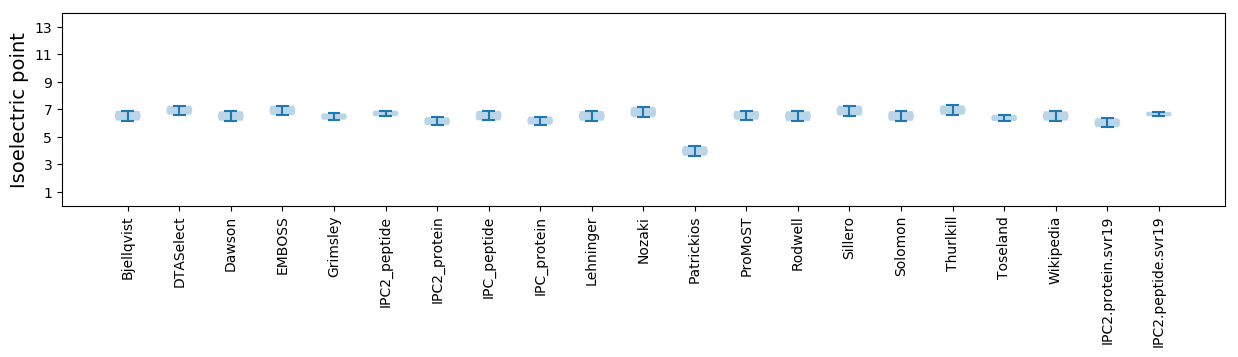
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJA3|A0A1L3KJA3_9VIRU Uncharacterized protein OS=Sanxia picorna-like virus 12 OX=1923369 PE=4 SV=1
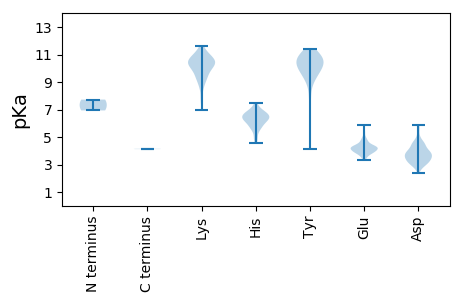
MM1 pKa = 6.93KK2 pKa = 9.76TSFVMPTEE10 pKa = 5.36DD11 pKa = 3.68YY12 pKa = 11.06QSLRR16 pKa = 11.84DD17 pKa = 3.71YY18 pKa = 10.37FARR21 pKa = 11.84PRR23 pKa = 11.84LIVTGALPTTRR34 pKa = 11.84ISFVNSEE41 pKa = 4.12VTPTVLFNTWFPGSLTRR58 pKa = 11.84LAGVHH63 pKa = 5.58GVRR66 pKa = 11.84FTLRR70 pKa = 11.84FTLTVAATPFQQALLCQSFQYY91 pKa = 9.79GTDD94 pKa = 3.2STIAAGSVIYY104 pKa = 10.33KK105 pKa = 10.42RR106 pKa = 11.84LGNSAMVTNLPHH118 pKa = 5.6VRR120 pKa = 11.84HH121 pKa = 6.6NITDD125 pKa = 3.43TTMTVFDD132 pKa = 4.49VPFLYY137 pKa = 10.69AKK139 pKa = 10.38DD140 pKa = 4.14YY141 pKa = 11.33LPLYY145 pKa = 10.04YY146 pKa = 10.41DD147 pKa = 4.71GSSSTNSLGQYY158 pKa = 9.01SANALLPYY166 pKa = 9.98RR167 pKa = 11.84AVAGANAPTFKK178 pKa = 10.63LFVSIHH184 pKa = 4.91EE185 pKa = 4.49LEE187 pKa = 5.38LIGSAPQVATTVVVQSGAGRR207 pKa = 11.84KK208 pKa = 8.78EE209 pKa = 3.99SLVGGNNPKK218 pKa = 10.07EE219 pKa = 4.09AEE221 pKa = 4.2KK222 pKa = 10.99KK223 pKa = 10.42SMKK226 pKa = 10.52LSDD229 pKa = 4.07RR230 pKa = 11.84LSQASKK236 pKa = 10.25VATIAAFVPALTGVAGPTAIAAEE259 pKa = 4.04AAARR263 pKa = 11.84VARR266 pKa = 11.84AFGYY270 pKa = 10.19SRR272 pKa = 11.84PQVEE276 pKa = 4.44DD277 pKa = 3.13QPTRR281 pKa = 11.84VYY283 pKa = 10.9RR284 pKa = 11.84NNYY287 pKa = 8.73CADD290 pKa = 3.83ANVDD294 pKa = 3.53IPSNSYY300 pKa = 10.5VLAPMQRR307 pKa = 11.84NEE309 pKa = 3.64VRR311 pKa = 11.84TDD313 pKa = 2.97AMAGGTDD320 pKa = 3.49VDD322 pKa = 4.46EE323 pKa = 4.42MALAYY328 pKa = 10.45VLEE331 pKa = 4.68KK332 pKa = 10.25PCQIFAGDD340 pKa = 4.27FSTTDD345 pKa = 3.12SVGTTLYY352 pKa = 10.94GSAVCPTNFWFRR364 pKa = 11.84SNSARR369 pKa = 11.84PGGNITLPSTAGLTTNAIYY388 pKa = 9.72PSTLCYY394 pKa = 9.18IGSMFRR400 pKa = 11.84YY401 pKa = 8.34WRR403 pKa = 11.84GGLKK407 pKa = 9.13FTFTFSKK414 pKa = 9.9TKK416 pKa = 9.25FHH418 pKa = 6.84GGRR421 pKa = 11.84VIVAFVPGLTDD432 pKa = 3.33NPSNSVLSNTIPTIEE447 pKa = 4.4NGGLPQPFSYY457 pKa = 11.1CEE459 pKa = 3.87VFDD462 pKa = 5.85LRR464 pKa = 11.84DD465 pKa = 3.2ADD467 pKa = 3.63TFEE470 pKa = 4.03FHH472 pKa = 6.43VPYY475 pKa = 10.6VSPTPYY481 pKa = 10.15TPVTASIGGITMTVLDD497 pKa = 4.63PLVSSGEE504 pKa = 4.07TSGTVDD510 pKa = 4.01YY511 pKa = 10.57IVEE514 pKa = 4.23VQAEE518 pKa = 3.98PDD520 pKa = 3.45FRR522 pKa = 11.84LGCPAPCMFTPASSTGTGVVFQQSGLGGVSDD553 pKa = 5.1LPDD556 pKa = 3.6NVDD559 pKa = 3.37EE560 pKa = 4.44YY561 pKa = 9.96TTGEE565 pKa = 4.4TILSLKK571 pKa = 8.33TLMMIPTYY579 pKa = 11.0VVDD582 pKa = 4.53SFDD585 pKa = 4.99AGTTSLWEE593 pKa = 3.74AWPYY597 pKa = 5.85WTRR600 pKa = 11.84SLWTMAVPMAPVTSKK615 pKa = 9.33TFAFSRR621 pKa = 11.84SGNIAACYY629 pKa = 9.64AFVVGSTEE637 pKa = 3.66HH638 pKa = 6.69HH639 pKa = 6.8VYY641 pKa = 10.75CPVTNGVYY649 pKa = 10.32ISASANEE656 pKa = 4.01RR657 pKa = 11.84DD658 pKa = 3.83GNAGAGAADD667 pKa = 3.67PRR669 pKa = 11.84SRR671 pKa = 11.84SVSGKK676 pKa = 9.28QRR678 pKa = 11.84VLTTDD683 pKa = 2.7NSLHH687 pKa = 6.27LRR689 pKa = 11.84VPLYY693 pKa = 10.98ARR695 pKa = 11.84VARR698 pKa = 11.84VPLASSSAAFGADD711 pKa = 2.36IGTTTFAGGGDD722 pKa = 3.84KK723 pKa = 11.03YY724 pKa = 10.84ISNYY728 pKa = 8.63NQILVRR734 pKa = 11.84NGTTAAIPFFYY745 pKa = 10.47GRR747 pKa = 11.84AAADD751 pKa = 3.82DD752 pKa = 3.99ARR754 pKa = 11.84CVGWLGPPPVVLFSSTVTFGPDD776 pKa = 2.9SPGDD780 pKa = 3.6FNFF783 pKa = 4.12
MM1 pKa = 6.93KK2 pKa = 9.76TSFVMPTEE10 pKa = 5.36DD11 pKa = 3.68YY12 pKa = 11.06QSLRR16 pKa = 11.84DD17 pKa = 3.71YY18 pKa = 10.37FARR21 pKa = 11.84PRR23 pKa = 11.84LIVTGALPTTRR34 pKa = 11.84ISFVNSEE41 pKa = 4.12VTPTVLFNTWFPGSLTRR58 pKa = 11.84LAGVHH63 pKa = 5.58GVRR66 pKa = 11.84FTLRR70 pKa = 11.84FTLTVAATPFQQALLCQSFQYY91 pKa = 9.79GTDD94 pKa = 3.2STIAAGSVIYY104 pKa = 10.33KK105 pKa = 10.42RR106 pKa = 11.84LGNSAMVTNLPHH118 pKa = 5.6VRR120 pKa = 11.84HH121 pKa = 6.6NITDD125 pKa = 3.43TTMTVFDD132 pKa = 4.49VPFLYY137 pKa = 10.69AKK139 pKa = 10.38DD140 pKa = 4.14YY141 pKa = 11.33LPLYY145 pKa = 10.04YY146 pKa = 10.41DD147 pKa = 4.71GSSSTNSLGQYY158 pKa = 9.01SANALLPYY166 pKa = 9.98RR167 pKa = 11.84AVAGANAPTFKK178 pKa = 10.63LFVSIHH184 pKa = 4.91EE185 pKa = 4.49LEE187 pKa = 5.38LIGSAPQVATTVVVQSGAGRR207 pKa = 11.84KK208 pKa = 8.78EE209 pKa = 3.99SLVGGNNPKK218 pKa = 10.07EE219 pKa = 4.09AEE221 pKa = 4.2KK222 pKa = 10.99KK223 pKa = 10.42SMKK226 pKa = 10.52LSDD229 pKa = 4.07RR230 pKa = 11.84LSQASKK236 pKa = 10.25VATIAAFVPALTGVAGPTAIAAEE259 pKa = 4.04AAARR263 pKa = 11.84VARR266 pKa = 11.84AFGYY270 pKa = 10.19SRR272 pKa = 11.84PQVEE276 pKa = 4.44DD277 pKa = 3.13QPTRR281 pKa = 11.84VYY283 pKa = 10.9RR284 pKa = 11.84NNYY287 pKa = 8.73CADD290 pKa = 3.83ANVDD294 pKa = 3.53IPSNSYY300 pKa = 10.5VLAPMQRR307 pKa = 11.84NEE309 pKa = 3.64VRR311 pKa = 11.84TDD313 pKa = 2.97AMAGGTDD320 pKa = 3.49VDD322 pKa = 4.46EE323 pKa = 4.42MALAYY328 pKa = 10.45VLEE331 pKa = 4.68KK332 pKa = 10.25PCQIFAGDD340 pKa = 4.27FSTTDD345 pKa = 3.12SVGTTLYY352 pKa = 10.94GSAVCPTNFWFRR364 pKa = 11.84SNSARR369 pKa = 11.84PGGNITLPSTAGLTTNAIYY388 pKa = 9.72PSTLCYY394 pKa = 9.18IGSMFRR400 pKa = 11.84YY401 pKa = 8.34WRR403 pKa = 11.84GGLKK407 pKa = 9.13FTFTFSKK414 pKa = 9.9TKK416 pKa = 9.25FHH418 pKa = 6.84GGRR421 pKa = 11.84VIVAFVPGLTDD432 pKa = 3.33NPSNSVLSNTIPTIEE447 pKa = 4.4NGGLPQPFSYY457 pKa = 11.1CEE459 pKa = 3.87VFDD462 pKa = 5.85LRR464 pKa = 11.84DD465 pKa = 3.2ADD467 pKa = 3.63TFEE470 pKa = 4.03FHH472 pKa = 6.43VPYY475 pKa = 10.6VSPTPYY481 pKa = 10.15TPVTASIGGITMTVLDD497 pKa = 4.63PLVSSGEE504 pKa = 4.07TSGTVDD510 pKa = 4.01YY511 pKa = 10.57IVEE514 pKa = 4.23VQAEE518 pKa = 3.98PDD520 pKa = 3.45FRR522 pKa = 11.84LGCPAPCMFTPASSTGTGVVFQQSGLGGVSDD553 pKa = 5.1LPDD556 pKa = 3.6NVDD559 pKa = 3.37EE560 pKa = 4.44YY561 pKa = 9.96TTGEE565 pKa = 4.4TILSLKK571 pKa = 8.33TLMMIPTYY579 pKa = 11.0VVDD582 pKa = 4.53SFDD585 pKa = 4.99AGTTSLWEE593 pKa = 3.74AWPYY597 pKa = 5.85WTRR600 pKa = 11.84SLWTMAVPMAPVTSKK615 pKa = 9.33TFAFSRR621 pKa = 11.84SGNIAACYY629 pKa = 9.64AFVVGSTEE637 pKa = 3.66HH638 pKa = 6.69HH639 pKa = 6.8VYY641 pKa = 10.75CPVTNGVYY649 pKa = 10.32ISASANEE656 pKa = 4.01RR657 pKa = 11.84DD658 pKa = 3.83GNAGAGAADD667 pKa = 3.67PRR669 pKa = 11.84SRR671 pKa = 11.84SVSGKK676 pKa = 9.28QRR678 pKa = 11.84VLTTDD683 pKa = 2.7NSLHH687 pKa = 6.27LRR689 pKa = 11.84VPLYY693 pKa = 10.98ARR695 pKa = 11.84VARR698 pKa = 11.84VPLASSSAAFGADD711 pKa = 2.36IGTTTFAGGGDD722 pKa = 3.84KK723 pKa = 11.03YY724 pKa = 10.84ISNYY728 pKa = 8.63NQILVRR734 pKa = 11.84NGTTAAIPFFYY745 pKa = 10.47GRR747 pKa = 11.84AAADD751 pKa = 3.82DD752 pKa = 3.99ARR754 pKa = 11.84CVGWLGPPPVVLFSSTVTFGPDD776 pKa = 2.9SPGDD780 pKa = 3.6FNFF783 pKa = 4.12
Molecular weight: 83.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJA3|A0A1L3KJA3_9VIRU Uncharacterized protein OS=Sanxia picorna-like virus 12 OX=1923369 PE=4 SV=1
MM1 pKa = 7.72SSINANAKK9 pKa = 9.0MYY11 pKa = 11.23ANGLVVKK18 pKa = 10.23SAAQARR24 pKa = 11.84IEE26 pKa = 4.21RR27 pKa = 11.84DD28 pKa = 2.72HH29 pKa = 6.28HH30 pKa = 4.58QQYY33 pKa = 9.45VAKK36 pKa = 10.05QRR38 pKa = 11.84YY39 pKa = 4.13TALRR43 pKa = 11.84KK44 pKa = 9.19EE45 pKa = 4.15IPKK48 pKa = 10.18KK49 pKa = 9.85LRR51 pKa = 11.84EE52 pKa = 4.05RR53 pKa = 11.84EE54 pKa = 3.82IEE56 pKa = 3.72DD57 pKa = 3.04ARR59 pKa = 11.84RR60 pKa = 11.84VKK62 pKa = 10.77RR63 pKa = 11.84MLNEE67 pKa = 4.54GEE69 pKa = 4.0SSEE72 pKa = 4.17YY73 pKa = 10.94GGIRR77 pKa = 11.84EE78 pKa = 4.17QGFASTIAVSAVGAAAAAAMYY99 pKa = 10.17SVNRR103 pKa = 11.84MRR105 pKa = 11.84KK106 pKa = 7.86EE107 pKa = 3.46VSGAVDD113 pKa = 3.24RR114 pKa = 11.84VDD116 pKa = 3.26EE117 pKa = 4.58ALASTSVSVVSTAHH131 pKa = 6.45LLVDD135 pKa = 4.26RR136 pKa = 11.84VNTTIEE142 pKa = 4.13GFANTAKK149 pKa = 10.64SILGAFWIVPTAILAHH165 pKa = 6.15YY166 pKa = 9.79IVGHH170 pKa = 6.21FSDD173 pKa = 4.66IPLLPVMASLFLAKK187 pKa = 10.62LFGDD191 pKa = 4.8EE192 pKa = 3.81IWNLLGKK199 pKa = 9.23WFAFQEE205 pKa = 4.24QSGEE209 pKa = 4.32GACSIGALIISAACAVCIPTRR230 pKa = 11.84SASGVAGEE238 pKa = 3.96IMKK241 pKa = 10.65RR242 pKa = 11.84MSSYY246 pKa = 10.57VRR248 pKa = 11.84AKK250 pKa = 10.62EE251 pKa = 3.86GFEE254 pKa = 4.91CFFKK258 pKa = 11.07DD259 pKa = 3.07ALKK262 pKa = 10.06YY263 pKa = 10.71AEE265 pKa = 4.23RR266 pKa = 11.84AVNAILSFFSEE277 pKa = 5.46KK278 pKa = 9.4EE279 pKa = 4.22VNWIGTSEE287 pKa = 4.12RR288 pKa = 11.84LVDD291 pKa = 4.0RR292 pKa = 11.84FCDD295 pKa = 3.16KK296 pKa = 10.41VTVFEE301 pKa = 4.41SLVRR305 pKa = 11.84TEE307 pKa = 3.82SDD309 pKa = 3.01KK310 pKa = 11.62VQLADD315 pKa = 3.84LLAAVSVQQEE325 pKa = 4.03ALGLKK330 pKa = 7.75MTVRR334 pKa = 11.84DD335 pKa = 3.98PQTLLRR341 pKa = 11.84IEE343 pKa = 4.64RR344 pKa = 11.84GLTRR348 pKa = 11.84LSLLLQPYY356 pKa = 9.36QGAITASRR364 pKa = 11.84NFRR367 pKa = 11.84PEE369 pKa = 3.38PTFICMYY376 pKa = 9.72GGSGVGKK383 pKa = 6.95TTLATKK389 pKa = 7.69MACTILVKK397 pKa = 10.66SGLCEE402 pKa = 3.95PDD404 pKa = 2.87EE405 pKa = 4.28ALRR408 pKa = 11.84NLWQKK413 pKa = 11.47GNTEE417 pKa = 3.85YY418 pKa = 10.73WNGYY422 pKa = 7.47VNQKK426 pKa = 9.86CLVMDD431 pKa = 4.3DD432 pKa = 4.47CFQVKK437 pKa = 8.72PVKK440 pKa = 10.87GEE442 pKa = 3.87TDD444 pKa = 3.29SEE446 pKa = 4.54YY447 pKa = 10.61MNVIRR452 pKa = 11.84MIGNWAYY459 pKa = 10.81ALNFADD465 pKa = 5.64LEE467 pKa = 4.84SKK469 pKa = 10.82GKK471 pKa = 10.47FYY473 pKa = 11.09FDD475 pKa = 3.49TPLVIGTTNCANIYY489 pKa = 10.76NMADD493 pKa = 3.24MVVTEE498 pKa = 4.22PAAVVRR504 pKa = 11.84RR505 pKa = 11.84IKK507 pKa = 10.22FPYY510 pKa = 9.49RR511 pKa = 11.84IEE513 pKa = 4.34VNPQYY518 pKa = 10.22WGEE521 pKa = 4.0EE522 pKa = 3.72KK523 pKa = 10.66QLDD526 pKa = 4.13YY527 pKa = 11.4KK528 pKa = 10.99KK529 pKa = 11.14LEE531 pKa = 4.33AEE533 pKa = 5.03FSANLDD539 pKa = 3.66AYY541 pKa = 10.02DD542 pKa = 3.6KK543 pKa = 10.88RR544 pKa = 11.84GEE546 pKa = 4.07RR547 pKa = 11.84SSLSASMDD555 pKa = 3.55AYY557 pKa = 9.9PWEE560 pKa = 3.79AWKK563 pKa = 10.39LVRR566 pKa = 11.84HH567 pKa = 6.26DD568 pKa = 4.33FSNPEE573 pKa = 3.57AGGPEE578 pKa = 3.75RR579 pKa = 11.84TVRR582 pKa = 11.84SLIDD586 pKa = 3.46EE587 pKa = 4.16VVEE590 pKa = 4.78RR591 pKa = 11.84IVSSRR596 pKa = 11.84EE597 pKa = 3.54AHH599 pKa = 5.99LQSVDD604 pKa = 2.99NLNRR608 pKa = 11.84FMRR611 pKa = 11.84NIGDD615 pKa = 3.47AMDD618 pKa = 4.53AGRR621 pKa = 11.84GVKK624 pKa = 9.47EE625 pKa = 3.77QGGLSEE631 pKa = 4.21ATSLQGDD638 pKa = 4.67FYY640 pKa = 10.22EE641 pKa = 4.9DD642 pKa = 3.63CVEE645 pKa = 4.21EE646 pKa = 4.33STSTVDD652 pKa = 2.94KK653 pKa = 11.1SAIEE657 pKa = 3.78RR658 pKa = 11.84ALRR661 pKa = 11.84IASGEE666 pKa = 3.98LDD668 pKa = 3.27VDD670 pKa = 3.93LDD672 pKa = 4.03EE673 pKa = 4.79IRR675 pKa = 11.84PVRR678 pKa = 11.84LTEE681 pKa = 4.01SVRR684 pKa = 11.84DD685 pKa = 3.83FVMDD689 pKa = 4.63GIPRR693 pKa = 11.84MSVGADD699 pKa = 3.33TPRR702 pKa = 11.84ASSFHH707 pKa = 6.6AHH709 pKa = 6.77INDD712 pKa = 3.54TQYY715 pKa = 11.08RR716 pKa = 11.84RR717 pKa = 11.84PNRR720 pKa = 11.84PSYY723 pKa = 9.74SFRR726 pKa = 11.84YY727 pKa = 10.2VIMKK731 pKa = 10.36RR732 pKa = 11.84LAQEE736 pKa = 4.0VYY738 pKa = 10.37KK739 pKa = 10.57RR740 pKa = 11.84IYY742 pKa = 9.95SWAEE746 pKa = 3.36NVRR749 pKa = 11.84KK750 pKa = 9.6FFTGLTAGGVIKK762 pKa = 10.21FAFCTLAATMVVGLAVGVFAAVRR785 pKa = 11.84YY786 pKa = 9.23IFKK789 pKa = 10.47AVKK792 pKa = 9.99SKK794 pKa = 10.68FAGTPRR800 pKa = 11.84GTPQSNVKK808 pKa = 10.08EE809 pKa = 4.29SAGPSRR815 pKa = 11.84KK816 pKa = 9.69ALFKK820 pKa = 10.38PKK822 pKa = 10.14SIKK825 pKa = 8.67TQSDD829 pKa = 3.14IQRR832 pKa = 11.84NRR834 pKa = 11.84VHH836 pKa = 6.73NNIFANTFKK845 pKa = 10.96LVVHH849 pKa = 6.92AEE851 pKa = 4.03EE852 pKa = 3.97QDD854 pKa = 3.8FVVGQIQFVQLHH866 pKa = 5.01VAMMPKK872 pKa = 9.97HH873 pKa = 6.17FRR875 pKa = 11.84RR876 pKa = 11.84QLEE879 pKa = 3.82EE880 pKa = 3.84RR881 pKa = 11.84LADD884 pKa = 3.81GGVKK888 pKa = 8.62EE889 pKa = 4.25TSLMSFVHH897 pKa = 5.94ATHH900 pKa = 5.76NQIVKK905 pKa = 9.57FKK907 pKa = 10.41VGEE910 pKa = 3.96FLACKK915 pKa = 9.61IVEE918 pKa = 4.32VEE920 pKa = 4.67DD921 pKa = 3.85SDD923 pKa = 4.59LLFMRR928 pKa = 11.84FPNGFISAASKK939 pKa = 8.76ITQHH943 pKa = 5.76FLKK946 pKa = 10.92AEE948 pKa = 4.32DD949 pKa = 3.93LNKK952 pKa = 10.0VIQAKK957 pKa = 9.24PAIRR961 pKa = 11.84LDD963 pKa = 3.32IAEE966 pKa = 5.84LDD968 pKa = 4.32LKK970 pKa = 9.94WQEE973 pKa = 3.95KK974 pKa = 10.56SKK976 pKa = 11.03GALISRR982 pKa = 11.84NTMLANTFNFLPNLASQAYY1001 pKa = 7.87EE1002 pKa = 3.61NSNVLEE1008 pKa = 4.03YY1009 pKa = 11.02QMPTEE1014 pKa = 4.49EE1015 pKa = 4.65GMCGAPLCISEE1026 pKa = 3.49NRR1028 pKa = 11.84YY1029 pKa = 10.18YY1030 pKa = 11.13GGACYY1035 pKa = 10.25LGMHH1039 pKa = 6.44VAGAPGWFSRR1049 pKa = 11.84VGYY1052 pKa = 10.25SNVVTQEE1059 pKa = 3.54QALAAVAEE1067 pKa = 4.62LEE1069 pKa = 4.4TVTDD1073 pKa = 3.79NFRR1076 pKa = 11.84EE1077 pKa = 4.14DD1078 pKa = 2.97AKK1080 pKa = 11.15KK1081 pKa = 10.36RR1082 pKa = 11.84GFPIVDD1088 pKa = 3.42VDD1090 pKa = 3.86PEE1092 pKa = 4.3EE1093 pKa = 4.31QSGLLTNSFITGSFTLIGRR1112 pKa = 11.84AEE1114 pKa = 4.27KK1115 pKa = 10.25PVGLNPSTKK1124 pKa = 10.41LILSPIGEE1132 pKa = 4.24EE1133 pKa = 4.17QPFGLNSLAPARR1145 pKa = 11.84LRR1147 pKa = 11.84PFMRR1151 pKa = 11.84DD1152 pKa = 3.21GEE1154 pKa = 4.57RR1155 pKa = 11.84VSPMSVGLSGYY1166 pKa = 7.55ATPLDD1171 pKa = 4.29YY1172 pKa = 10.93RR1173 pKa = 11.84PLPNMDD1179 pKa = 5.6AIVSMATKK1187 pKa = 10.02PFRR1190 pKa = 11.84LASIGDD1196 pKa = 3.46YY1197 pKa = 10.69RR1198 pKa = 11.84GILTKK1203 pKa = 10.41RR1204 pKa = 11.84EE1205 pKa = 3.97AVEE1208 pKa = 4.76GIEE1211 pKa = 4.31GMKK1214 pKa = 10.19LKK1216 pKa = 10.5PIARR1220 pKa = 11.84GKK1222 pKa = 9.71SPGYY1226 pKa = 9.49PFVHH1230 pKa = 6.46EE1231 pKa = 4.18VRR1233 pKa = 11.84NGKK1236 pKa = 9.33RR1237 pKa = 11.84EE1238 pKa = 3.84YY1239 pKa = 10.41FGNDD1243 pKa = 2.9GPFEE1247 pKa = 4.23FEE1249 pKa = 4.4SPQCVKK1255 pKa = 10.72LLARR1259 pKa = 11.84VDD1261 pKa = 3.82EE1262 pKa = 5.03VIANAKK1268 pKa = 9.78EE1269 pKa = 4.21GVRR1272 pKa = 11.84LCHH1275 pKa = 6.72IFTDD1279 pKa = 4.35FLKK1282 pKa = 11.09DD1283 pKa = 3.04EE1284 pKa = 4.3TRR1286 pKa = 11.84PLRR1289 pKa = 11.84KK1290 pKa = 9.48VEE1292 pKa = 3.95QGEE1295 pKa = 4.26TRR1297 pKa = 11.84VISGAPLDD1305 pKa = 4.38LVIATRR1311 pKa = 11.84MYY1313 pKa = 10.12FGAWMASMFRR1323 pKa = 11.84HH1324 pKa = 5.5CVVSGVAPGINPYY1337 pKa = 9.99NEE1339 pKa = 3.97WYY1341 pKa = 10.68LLASRR1346 pKa = 11.84LSSKK1350 pKa = 10.54GEE1352 pKa = 3.91KK1353 pKa = 10.59VFDD1356 pKa = 3.98GDD1358 pKa = 4.58FKK1360 pKa = 11.62AFDD1363 pKa = 3.94KK1364 pKa = 11.4SEE1366 pKa = 3.93QPYY1369 pKa = 9.19VHH1371 pKa = 6.4YY1372 pKa = 10.71AILDD1376 pKa = 4.51FINHH1380 pKa = 6.39WYY1382 pKa = 10.59GGTEE1386 pKa = 3.33EE1387 pKa = 4.35DD1388 pKa = 3.55EE1389 pKa = 4.36RR1390 pKa = 11.84VRR1392 pKa = 11.84GILWLEE1398 pKa = 4.1LVHH1401 pKa = 6.67SRR1403 pKa = 11.84HH1404 pKa = 6.1LGGDD1408 pKa = 3.53GVVQDD1413 pKa = 5.82IVYY1416 pKa = 9.29QWSKK1420 pKa = 11.34SMPSGHH1426 pKa = 7.48PLTTPVNSMYY1436 pKa = 11.22SLITLTGCYY1445 pKa = 10.14AKK1447 pKa = 10.04ATGDD1451 pKa = 3.68NEE1453 pKa = 4.95SMWEE1457 pKa = 3.81HH1458 pKa = 6.57AFINTFGDD1466 pKa = 3.87DD1467 pKa = 3.79NITNVSDD1474 pKa = 3.49KK1475 pKa = 10.7VAEE1478 pKa = 4.21VFNQVTVARR1487 pKa = 11.84DD1488 pKa = 3.25MDD1490 pKa = 4.11EE1491 pKa = 5.3LFGLTYY1497 pKa = 9.9TAGSKK1502 pKa = 9.15TGEE1505 pKa = 4.12LVPYY1509 pKa = 9.33TDD1511 pKa = 5.26LGSCTFLKK1519 pKa = 10.47RR1520 pKa = 11.84SFVRR1524 pKa = 11.84DD1525 pKa = 3.41VAGSGGWIAPIDD1537 pKa = 4.0ITSHH1541 pKa = 6.99LYY1543 pKa = 9.16SCYY1546 pKa = 9.56YY1547 pKa = 9.73YY1548 pKa = 10.95KK1549 pKa = 10.85NKK1551 pKa = 9.72RR1552 pKa = 11.84DD1553 pKa = 3.76PNADD1557 pKa = 3.04MAHH1560 pKa = 6.34NLEE1563 pKa = 4.2EE1564 pKa = 4.21MLGEE1568 pKa = 4.32LSLHH1572 pKa = 7.18GIQVWEE1578 pKa = 4.31QYY1580 pKa = 11.0SGIAFEE1586 pKa = 4.84VLGHH1590 pKa = 5.77MGRR1593 pKa = 11.84VPRR1596 pKa = 11.84YY1597 pKa = 5.57MTRR1600 pKa = 11.84EE1601 pKa = 4.69GYY1603 pKa = 10.34RR1604 pKa = 11.84SMMRR1608 pKa = 11.84QRR1610 pKa = 11.84LDD1612 pKa = 2.6AWFF1615 pKa = 4.14
MM1 pKa = 7.72SSINANAKK9 pKa = 9.0MYY11 pKa = 11.23ANGLVVKK18 pKa = 10.23SAAQARR24 pKa = 11.84IEE26 pKa = 4.21RR27 pKa = 11.84DD28 pKa = 2.72HH29 pKa = 6.28HH30 pKa = 4.58QQYY33 pKa = 9.45VAKK36 pKa = 10.05QRR38 pKa = 11.84YY39 pKa = 4.13TALRR43 pKa = 11.84KK44 pKa = 9.19EE45 pKa = 4.15IPKK48 pKa = 10.18KK49 pKa = 9.85LRR51 pKa = 11.84EE52 pKa = 4.05RR53 pKa = 11.84EE54 pKa = 3.82IEE56 pKa = 3.72DD57 pKa = 3.04ARR59 pKa = 11.84RR60 pKa = 11.84VKK62 pKa = 10.77RR63 pKa = 11.84MLNEE67 pKa = 4.54GEE69 pKa = 4.0SSEE72 pKa = 4.17YY73 pKa = 10.94GGIRR77 pKa = 11.84EE78 pKa = 4.17QGFASTIAVSAVGAAAAAAMYY99 pKa = 10.17SVNRR103 pKa = 11.84MRR105 pKa = 11.84KK106 pKa = 7.86EE107 pKa = 3.46VSGAVDD113 pKa = 3.24RR114 pKa = 11.84VDD116 pKa = 3.26EE117 pKa = 4.58ALASTSVSVVSTAHH131 pKa = 6.45LLVDD135 pKa = 4.26RR136 pKa = 11.84VNTTIEE142 pKa = 4.13GFANTAKK149 pKa = 10.64SILGAFWIVPTAILAHH165 pKa = 6.15YY166 pKa = 9.79IVGHH170 pKa = 6.21FSDD173 pKa = 4.66IPLLPVMASLFLAKK187 pKa = 10.62LFGDD191 pKa = 4.8EE192 pKa = 3.81IWNLLGKK199 pKa = 9.23WFAFQEE205 pKa = 4.24QSGEE209 pKa = 4.32GACSIGALIISAACAVCIPTRR230 pKa = 11.84SASGVAGEE238 pKa = 3.96IMKK241 pKa = 10.65RR242 pKa = 11.84MSSYY246 pKa = 10.57VRR248 pKa = 11.84AKK250 pKa = 10.62EE251 pKa = 3.86GFEE254 pKa = 4.91CFFKK258 pKa = 11.07DD259 pKa = 3.07ALKK262 pKa = 10.06YY263 pKa = 10.71AEE265 pKa = 4.23RR266 pKa = 11.84AVNAILSFFSEE277 pKa = 5.46KK278 pKa = 9.4EE279 pKa = 4.22VNWIGTSEE287 pKa = 4.12RR288 pKa = 11.84LVDD291 pKa = 4.0RR292 pKa = 11.84FCDD295 pKa = 3.16KK296 pKa = 10.41VTVFEE301 pKa = 4.41SLVRR305 pKa = 11.84TEE307 pKa = 3.82SDD309 pKa = 3.01KK310 pKa = 11.62VQLADD315 pKa = 3.84LLAAVSVQQEE325 pKa = 4.03ALGLKK330 pKa = 7.75MTVRR334 pKa = 11.84DD335 pKa = 3.98PQTLLRR341 pKa = 11.84IEE343 pKa = 4.64RR344 pKa = 11.84GLTRR348 pKa = 11.84LSLLLQPYY356 pKa = 9.36QGAITASRR364 pKa = 11.84NFRR367 pKa = 11.84PEE369 pKa = 3.38PTFICMYY376 pKa = 9.72GGSGVGKK383 pKa = 6.95TTLATKK389 pKa = 7.69MACTILVKK397 pKa = 10.66SGLCEE402 pKa = 3.95PDD404 pKa = 2.87EE405 pKa = 4.28ALRR408 pKa = 11.84NLWQKK413 pKa = 11.47GNTEE417 pKa = 3.85YY418 pKa = 10.73WNGYY422 pKa = 7.47VNQKK426 pKa = 9.86CLVMDD431 pKa = 4.3DD432 pKa = 4.47CFQVKK437 pKa = 8.72PVKK440 pKa = 10.87GEE442 pKa = 3.87TDD444 pKa = 3.29SEE446 pKa = 4.54YY447 pKa = 10.61MNVIRR452 pKa = 11.84MIGNWAYY459 pKa = 10.81ALNFADD465 pKa = 5.64LEE467 pKa = 4.84SKK469 pKa = 10.82GKK471 pKa = 10.47FYY473 pKa = 11.09FDD475 pKa = 3.49TPLVIGTTNCANIYY489 pKa = 10.76NMADD493 pKa = 3.24MVVTEE498 pKa = 4.22PAAVVRR504 pKa = 11.84RR505 pKa = 11.84IKK507 pKa = 10.22FPYY510 pKa = 9.49RR511 pKa = 11.84IEE513 pKa = 4.34VNPQYY518 pKa = 10.22WGEE521 pKa = 4.0EE522 pKa = 3.72KK523 pKa = 10.66QLDD526 pKa = 4.13YY527 pKa = 11.4KK528 pKa = 10.99KK529 pKa = 11.14LEE531 pKa = 4.33AEE533 pKa = 5.03FSANLDD539 pKa = 3.66AYY541 pKa = 10.02DD542 pKa = 3.6KK543 pKa = 10.88RR544 pKa = 11.84GEE546 pKa = 4.07RR547 pKa = 11.84SSLSASMDD555 pKa = 3.55AYY557 pKa = 9.9PWEE560 pKa = 3.79AWKK563 pKa = 10.39LVRR566 pKa = 11.84HH567 pKa = 6.26DD568 pKa = 4.33FSNPEE573 pKa = 3.57AGGPEE578 pKa = 3.75RR579 pKa = 11.84TVRR582 pKa = 11.84SLIDD586 pKa = 3.46EE587 pKa = 4.16VVEE590 pKa = 4.78RR591 pKa = 11.84IVSSRR596 pKa = 11.84EE597 pKa = 3.54AHH599 pKa = 5.99LQSVDD604 pKa = 2.99NLNRR608 pKa = 11.84FMRR611 pKa = 11.84NIGDD615 pKa = 3.47AMDD618 pKa = 4.53AGRR621 pKa = 11.84GVKK624 pKa = 9.47EE625 pKa = 3.77QGGLSEE631 pKa = 4.21ATSLQGDD638 pKa = 4.67FYY640 pKa = 10.22EE641 pKa = 4.9DD642 pKa = 3.63CVEE645 pKa = 4.21EE646 pKa = 4.33STSTVDD652 pKa = 2.94KK653 pKa = 11.1SAIEE657 pKa = 3.78RR658 pKa = 11.84ALRR661 pKa = 11.84IASGEE666 pKa = 3.98LDD668 pKa = 3.27VDD670 pKa = 3.93LDD672 pKa = 4.03EE673 pKa = 4.79IRR675 pKa = 11.84PVRR678 pKa = 11.84LTEE681 pKa = 4.01SVRR684 pKa = 11.84DD685 pKa = 3.83FVMDD689 pKa = 4.63GIPRR693 pKa = 11.84MSVGADD699 pKa = 3.33TPRR702 pKa = 11.84ASSFHH707 pKa = 6.6AHH709 pKa = 6.77INDD712 pKa = 3.54TQYY715 pKa = 11.08RR716 pKa = 11.84RR717 pKa = 11.84PNRR720 pKa = 11.84PSYY723 pKa = 9.74SFRR726 pKa = 11.84YY727 pKa = 10.2VIMKK731 pKa = 10.36RR732 pKa = 11.84LAQEE736 pKa = 4.0VYY738 pKa = 10.37KK739 pKa = 10.57RR740 pKa = 11.84IYY742 pKa = 9.95SWAEE746 pKa = 3.36NVRR749 pKa = 11.84KK750 pKa = 9.6FFTGLTAGGVIKK762 pKa = 10.21FAFCTLAATMVVGLAVGVFAAVRR785 pKa = 11.84YY786 pKa = 9.23IFKK789 pKa = 10.47AVKK792 pKa = 9.99SKK794 pKa = 10.68FAGTPRR800 pKa = 11.84GTPQSNVKK808 pKa = 10.08EE809 pKa = 4.29SAGPSRR815 pKa = 11.84KK816 pKa = 9.69ALFKK820 pKa = 10.38PKK822 pKa = 10.14SIKK825 pKa = 8.67TQSDD829 pKa = 3.14IQRR832 pKa = 11.84NRR834 pKa = 11.84VHH836 pKa = 6.73NNIFANTFKK845 pKa = 10.96LVVHH849 pKa = 6.92AEE851 pKa = 4.03EE852 pKa = 3.97QDD854 pKa = 3.8FVVGQIQFVQLHH866 pKa = 5.01VAMMPKK872 pKa = 9.97HH873 pKa = 6.17FRR875 pKa = 11.84RR876 pKa = 11.84QLEE879 pKa = 3.82EE880 pKa = 3.84RR881 pKa = 11.84LADD884 pKa = 3.81GGVKK888 pKa = 8.62EE889 pKa = 4.25TSLMSFVHH897 pKa = 5.94ATHH900 pKa = 5.76NQIVKK905 pKa = 9.57FKK907 pKa = 10.41VGEE910 pKa = 3.96FLACKK915 pKa = 9.61IVEE918 pKa = 4.32VEE920 pKa = 4.67DD921 pKa = 3.85SDD923 pKa = 4.59LLFMRR928 pKa = 11.84FPNGFISAASKK939 pKa = 8.76ITQHH943 pKa = 5.76FLKK946 pKa = 10.92AEE948 pKa = 4.32DD949 pKa = 3.93LNKK952 pKa = 10.0VIQAKK957 pKa = 9.24PAIRR961 pKa = 11.84LDD963 pKa = 3.32IAEE966 pKa = 5.84LDD968 pKa = 4.32LKK970 pKa = 9.94WQEE973 pKa = 3.95KK974 pKa = 10.56SKK976 pKa = 11.03GALISRR982 pKa = 11.84NTMLANTFNFLPNLASQAYY1001 pKa = 7.87EE1002 pKa = 3.61NSNVLEE1008 pKa = 4.03YY1009 pKa = 11.02QMPTEE1014 pKa = 4.49EE1015 pKa = 4.65GMCGAPLCISEE1026 pKa = 3.49NRR1028 pKa = 11.84YY1029 pKa = 10.18YY1030 pKa = 11.13GGACYY1035 pKa = 10.25LGMHH1039 pKa = 6.44VAGAPGWFSRR1049 pKa = 11.84VGYY1052 pKa = 10.25SNVVTQEE1059 pKa = 3.54QALAAVAEE1067 pKa = 4.62LEE1069 pKa = 4.4TVTDD1073 pKa = 3.79NFRR1076 pKa = 11.84EE1077 pKa = 4.14DD1078 pKa = 2.97AKK1080 pKa = 11.15KK1081 pKa = 10.36RR1082 pKa = 11.84GFPIVDD1088 pKa = 3.42VDD1090 pKa = 3.86PEE1092 pKa = 4.3EE1093 pKa = 4.31QSGLLTNSFITGSFTLIGRR1112 pKa = 11.84AEE1114 pKa = 4.27KK1115 pKa = 10.25PVGLNPSTKK1124 pKa = 10.41LILSPIGEE1132 pKa = 4.24EE1133 pKa = 4.17QPFGLNSLAPARR1145 pKa = 11.84LRR1147 pKa = 11.84PFMRR1151 pKa = 11.84DD1152 pKa = 3.21GEE1154 pKa = 4.57RR1155 pKa = 11.84VSPMSVGLSGYY1166 pKa = 7.55ATPLDD1171 pKa = 4.29YY1172 pKa = 10.93RR1173 pKa = 11.84PLPNMDD1179 pKa = 5.6AIVSMATKK1187 pKa = 10.02PFRR1190 pKa = 11.84LASIGDD1196 pKa = 3.46YY1197 pKa = 10.69RR1198 pKa = 11.84GILTKK1203 pKa = 10.41RR1204 pKa = 11.84EE1205 pKa = 3.97AVEE1208 pKa = 4.76GIEE1211 pKa = 4.31GMKK1214 pKa = 10.19LKK1216 pKa = 10.5PIARR1220 pKa = 11.84GKK1222 pKa = 9.71SPGYY1226 pKa = 9.49PFVHH1230 pKa = 6.46EE1231 pKa = 4.18VRR1233 pKa = 11.84NGKK1236 pKa = 9.33RR1237 pKa = 11.84EE1238 pKa = 3.84YY1239 pKa = 10.41FGNDD1243 pKa = 2.9GPFEE1247 pKa = 4.23FEE1249 pKa = 4.4SPQCVKK1255 pKa = 10.72LLARR1259 pKa = 11.84VDD1261 pKa = 3.82EE1262 pKa = 5.03VIANAKK1268 pKa = 9.78EE1269 pKa = 4.21GVRR1272 pKa = 11.84LCHH1275 pKa = 6.72IFTDD1279 pKa = 4.35FLKK1282 pKa = 11.09DD1283 pKa = 3.04EE1284 pKa = 4.3TRR1286 pKa = 11.84PLRR1289 pKa = 11.84KK1290 pKa = 9.48VEE1292 pKa = 3.95QGEE1295 pKa = 4.26TRR1297 pKa = 11.84VISGAPLDD1305 pKa = 4.38LVIATRR1311 pKa = 11.84MYY1313 pKa = 10.12FGAWMASMFRR1323 pKa = 11.84HH1324 pKa = 5.5CVVSGVAPGINPYY1337 pKa = 9.99NEE1339 pKa = 3.97WYY1341 pKa = 10.68LLASRR1346 pKa = 11.84LSSKK1350 pKa = 10.54GEE1352 pKa = 3.91KK1353 pKa = 10.59VFDD1356 pKa = 3.98GDD1358 pKa = 4.58FKK1360 pKa = 11.62AFDD1363 pKa = 3.94KK1364 pKa = 11.4SEE1366 pKa = 3.93QPYY1369 pKa = 9.19VHH1371 pKa = 6.4YY1372 pKa = 10.71AILDD1376 pKa = 4.51FINHH1380 pKa = 6.39WYY1382 pKa = 10.59GGTEE1386 pKa = 3.33EE1387 pKa = 4.35DD1388 pKa = 3.55EE1389 pKa = 4.36RR1390 pKa = 11.84VRR1392 pKa = 11.84GILWLEE1398 pKa = 4.1LVHH1401 pKa = 6.67SRR1403 pKa = 11.84HH1404 pKa = 6.1LGGDD1408 pKa = 3.53GVVQDD1413 pKa = 5.82IVYY1416 pKa = 9.29QWSKK1420 pKa = 11.34SMPSGHH1426 pKa = 7.48PLTTPVNSMYY1436 pKa = 11.22SLITLTGCYY1445 pKa = 10.14AKK1447 pKa = 10.04ATGDD1451 pKa = 3.68NEE1453 pKa = 4.95SMWEE1457 pKa = 3.81HH1458 pKa = 6.57AFINTFGDD1466 pKa = 3.87DD1467 pKa = 3.79NITNVSDD1474 pKa = 3.49KK1475 pKa = 10.7VAEE1478 pKa = 4.21VFNQVTVARR1487 pKa = 11.84DD1488 pKa = 3.25MDD1490 pKa = 4.11EE1491 pKa = 5.3LFGLTYY1497 pKa = 9.9TAGSKK1502 pKa = 9.15TGEE1505 pKa = 4.12LVPYY1509 pKa = 9.33TDD1511 pKa = 5.26LGSCTFLKK1519 pKa = 10.47RR1520 pKa = 11.84SFVRR1524 pKa = 11.84DD1525 pKa = 3.41VAGSGGWIAPIDD1537 pKa = 4.0ITSHH1541 pKa = 6.99LYY1543 pKa = 9.16SCYY1546 pKa = 9.56YY1547 pKa = 9.73YY1548 pKa = 10.95KK1549 pKa = 10.85NKK1551 pKa = 9.72RR1552 pKa = 11.84DD1553 pKa = 3.76PNADD1557 pKa = 3.04MAHH1560 pKa = 6.34NLEE1563 pKa = 4.2EE1564 pKa = 4.21MLGEE1568 pKa = 4.32LSLHH1572 pKa = 7.18GIQVWEE1578 pKa = 4.31QYY1580 pKa = 11.0SGIAFEE1586 pKa = 4.84VLGHH1590 pKa = 5.77MGRR1593 pKa = 11.84VPRR1596 pKa = 11.84YY1597 pKa = 5.57MTRR1600 pKa = 11.84EE1601 pKa = 4.69GYY1603 pKa = 10.34RR1604 pKa = 11.84SMMRR1608 pKa = 11.84QRR1610 pKa = 11.84LDD1612 pKa = 2.6AWFF1615 pKa = 4.14
Molecular weight: 180.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2398 |
783 |
1615 |
1199.0 |
132.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.341 ± 0.374 | 1.418 ± 0.006 |
4.837 ± 0.056 | 5.713 ± 1.388 |
5.129 ± 0.245 | 7.59 ± 0.292 |
1.626 ± 0.238 | 4.587 ± 0.506 |
4.337 ± 1.019 | 7.631 ± 0.304 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.669 ± 0.377 | 4.128 ± 0.043 |
4.879 ± 0.945 | 2.877 ± 0.225 |
5.922 ± 0.534 | 7.548 ± 0.568 |
6.547 ± 1.771 | 8.215 ± 0.299 |
1.251 ± 0.115 | 3.753 ± 0.231 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
