
Mycobacterium marinum (strain ATCC BAA-535 / M)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium marinum
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
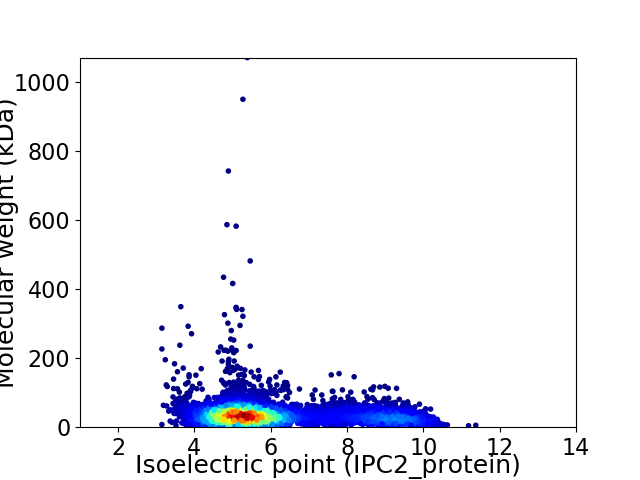
Virtual 2D-PAGE plot for 5418 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2HN11|B2HN11_MYCMM PE-PGRS family protein OS=Mycobacterium marinum (strain ATCC BAA-535 / M) OX=216594 GN=MMAR_5555 PE=4 SV=1
MM1 pKa = 7.59SYY3 pKa = 11.01VIATPDD9 pKa = 3.37MLAAAASDD17 pKa = 3.52LTNIGSTLGAANSAALARR35 pKa = 11.84TTGLLAAAEE44 pKa = 4.79DD45 pKa = 4.3EE46 pKa = 4.57VSTAIASLFSASAQEE61 pKa = 4.05YY62 pKa = 10.01QMVSTRR68 pKa = 11.84AAHH71 pKa = 5.78LHH73 pKa = 5.9AEE75 pKa = 4.74FVHH78 pKa = 6.04SLTRR82 pKa = 11.84AGAAYY87 pKa = 9.83AAAEE91 pKa = 4.21AVNASPLQTLEE102 pKa = 3.95QNVLAAINAPSQALTARR119 pKa = 11.84PLFGDD124 pKa = 4.35GADD127 pKa = 3.98GAPGTGQNGGDD138 pKa = 3.93GGWLIGNGGNGGSGAPGGNGGAGGSAGLWGRR169 pKa = 11.84GGNGGAGGTGVAGGAGGNGGSGGANGLLGGGWGPFGGASGGAGGAGGAGGIGVAGGLGGDD229 pKa = 4.35GGAGGAGGAGGANSQLLSLWGSAGAGGAGGIGGAGGAGTDD269 pKa = 4.12GSAGGIGADD278 pKa = 3.99GGAGGHH284 pKa = 6.27GGVGGAGGANNAWLGGAAGSGGTGGSGGTGGLGGAGGSGIGYY326 pKa = 7.74STGGGAGGAGGAGGDD341 pKa = 3.83AGTGGAAGATAFLGSAGVAGSAGNGGHH368 pKa = 6.67GGNGGSGADD377 pKa = 3.66GDD379 pKa = 3.97IGVAGSGAGADD390 pKa = 3.98GGNGFAGGTGGDD402 pKa = 3.91GGTGGNTTGGSNGSGGNAGHH422 pKa = 7.08GGNAGDD428 pKa = 4.06GAAGNTGVGFATKK441 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH477 pKa = 6.97GGNGGNGADD486 pKa = 4.09GDD488 pKa = 4.0IGVAGIGAGADD499 pKa = 3.56GGNGFAGGTGGDD511 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH531 pKa = 7.13GGNAGNGAAGNTGVGFATKK550 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH586 pKa = 6.97GGNGGNGADD595 pKa = 4.09GDD597 pKa = 4.0IGVAGIGAGADD608 pKa = 3.56GGNGFAGGTGGDD620 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH640 pKa = 7.13GGNAGNGAAGNTGVGFATKK659 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH695 pKa = 6.97GGNGGNGADD704 pKa = 4.09GDD706 pKa = 4.0IGVAGIGAGADD717 pKa = 3.56GGNGFAGGTGGDD729 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH749 pKa = 7.13GGNAGNGAAGNTGVGFATKK768 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH804 pKa = 6.97GGNGGNGADD813 pKa = 4.09GDD815 pKa = 4.0IGVAGIGAGADD826 pKa = 3.56GGNGFAGGTGGDD838 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH858 pKa = 7.13GGNAGNGADD867 pKa = 4.16GNVGDD872 pKa = 4.82YY873 pKa = 10.21YY874 pKa = 11.59VSGGTGGMGGDD885 pKa = 3.83GGDD888 pKa = 3.46AGTAGLAGTGGSGGTVGAYY907 pKa = 9.57SAGGAAGNGGTGGAGAQGAPAQFGSTGFAGGDD939 pKa = 3.31GGAGGDD945 pKa = 3.8GGTGGGGNGGTNGNGGHH962 pKa = 6.85GGTGGAGGQGGVGYY976 pKa = 9.23TDD978 pKa = 4.87DD979 pKa = 4.24GSNNATTGGDD989 pKa = 3.65GGDD992 pKa = 3.94GGQGGQGGVPGATGTGGTVGMAGNGGAGGAGGQGGDD1028 pKa = 3.43TTTSGNSAGFGGQGGQGGQGGAGLSGLTGGDD1059 pKa = 4.1GYY1061 pKa = 11.39DD1062 pKa = 3.8GGQGGAGGTGGAGGDD1077 pKa = 3.84AVSGAYY1083 pKa = 9.57NGHH1086 pKa = 6.59GGAGGDD1092 pKa = 4.02GGAGGQGGTGHH1103 pKa = 7.56ADD1105 pKa = 3.3DD1106 pKa = 5.27GSNNAGAGGNGGMGGQGGQGGGAGAGAGAGSGAGSAGTGGDD1147 pKa = 3.6GGTGGDD1153 pKa = 3.78GGATTTIGHH1162 pKa = 5.6TAGAGGVGGQGGDD1175 pKa = 3.76GGDD1178 pKa = 3.88GLADD1182 pKa = 3.64AASSGGKK1189 pKa = 9.99GGAGGLGGNGGDD1201 pKa = 3.83AVSGAYY1207 pKa = 9.75NGQGGMGGQGGTGGQGGDD1225 pKa = 4.05AGPEE1229 pKa = 4.09GNGGNGGNGGDD1240 pKa = 4.07GGTAGAAGTGGLGGTAGTAGAGGTGGGGGQGGDD1273 pKa = 3.27TTTAGHH1279 pKa = 5.39TAGWGGNGGYY1289 pKa = 10.11GGQGGGGAAGPAGGSGSDD1307 pKa = 3.77GGQGGQGGAGGTGGDD1322 pKa = 3.67AVAGATNGSGGTGGYY1337 pKa = 9.65GGQGGQGGAGGTDD1350 pKa = 3.2DD1351 pKa = 5.4GGDD1354 pKa = 3.41NAGTGGAGGTGGTGGAAGATGSGGTVDD1381 pKa = 4.38LAFPGTAGHH1390 pKa = 6.65GGDD1393 pKa = 4.05GGAGGGGGATTIVGHH1408 pKa = 5.14TAGTGGSGGQGGEE1421 pKa = 3.99GGAAADD1427 pKa = 3.82TGVPGAAGSTGGFGGTGGAGGAGGDD1452 pKa = 3.73AVAGATNGSGGAGGQGGAGAKK1473 pKa = 9.92GGQGYY1478 pKa = 10.67DD1479 pKa = 3.9DD1480 pKa = 5.81DD1481 pKa = 4.72GANNAGVGGNGGEE1494 pKa = 4.41GGTGGSGGAAGTGGDD1509 pKa = 3.95GGTAGAAGTGGTGGDD1524 pKa = 3.73GGKK1527 pKa = 10.64GGATTISGHH1536 pKa = 5.23HH1537 pKa = 6.56AGYY1540 pKa = 10.29GGNAGGGGQGADD1552 pKa = 3.71GAAGPAGGNGYY1563 pKa = 10.62DD1564 pKa = 4.03GGQGGAGGTGGQGGDD1579 pKa = 3.37AVAGATNGYY1588 pKa = 9.06GGQGGIGGTGGQGGGGGADD1607 pKa = 3.93DD1608 pKa = 5.2GSDD1611 pKa = 3.21NAGAGGSGGAGGLGGQSGAASTGGIVGTGGTGFAGDD1647 pKa = 4.43GGDD1650 pKa = 4.24GGAGGQGGATAVSGHH1665 pKa = 5.23TAGAGGTGGTGGTGGDD1681 pKa = 4.51GIDD1684 pKa = 4.33GPADD1688 pKa = 3.73RR1689 pKa = 11.84PGGRR1693 pKa = 11.84GGQGGHH1699 pKa = 6.5GGDD1702 pKa = 3.75GGSGGSAVAGATNGSGGAGGAGGTGGQGGNGYY1734 pKa = 10.56DD1735 pKa = 4.97DD1736 pKa = 5.59DD1737 pKa = 6.57GSDD1740 pKa = 3.29AAGAGGAGGDD1750 pKa = 3.72GGVGGTAGAVGSGGSGGTAGTAGTGGDD1777 pKa = 3.95GGDD1780 pKa = 3.74GGKK1783 pKa = 10.65GGATTTIGSSAGSGGDD1799 pKa = 3.41GGNGGLGGAGAIGPAGADD1817 pKa = 3.45GSTGGQGGAGGQGGDD1832 pKa = 3.98GGDD1835 pKa = 3.56AVAGATNGLGGNGGDD1850 pKa = 4.29GGSGGQGGAGGVDD1863 pKa = 3.6DD1864 pKa = 5.46GGGAGTGGQGGEE1876 pKa = 4.31GGQAGDD1882 pKa = 4.15GGVSGSGGLFGGGGDD1897 pKa = 4.27GGHH1900 pKa = 6.93GGQGGLGGTGGDD1912 pKa = 3.59SALGTGGDD1920 pKa = 4.15GGQGGQGGTGGTGGYY1935 pKa = 10.1GAFEE1939 pKa = 4.57GGFGGDD1945 pKa = 3.77GGDD1948 pKa = 3.79GGTGGTGGVGQSGLPGTGGTIGIAGVPGSLPSGSTGGAGGDD1989 pKa = 3.94GGDD1992 pKa = 4.58GGDD1995 pKa = 4.59GGPGSPP2001 pKa = 4.79
MM1 pKa = 7.59SYY3 pKa = 11.01VIATPDD9 pKa = 3.37MLAAAASDD17 pKa = 3.52LTNIGSTLGAANSAALARR35 pKa = 11.84TTGLLAAAEE44 pKa = 4.79DD45 pKa = 4.3EE46 pKa = 4.57VSTAIASLFSASAQEE61 pKa = 4.05YY62 pKa = 10.01QMVSTRR68 pKa = 11.84AAHH71 pKa = 5.78LHH73 pKa = 5.9AEE75 pKa = 4.74FVHH78 pKa = 6.04SLTRR82 pKa = 11.84AGAAYY87 pKa = 9.83AAAEE91 pKa = 4.21AVNASPLQTLEE102 pKa = 3.95QNVLAAINAPSQALTARR119 pKa = 11.84PLFGDD124 pKa = 4.35GADD127 pKa = 3.98GAPGTGQNGGDD138 pKa = 3.93GGWLIGNGGNGGSGAPGGNGGAGGSAGLWGRR169 pKa = 11.84GGNGGAGGTGVAGGAGGNGGSGGANGLLGGGWGPFGGASGGAGGAGGAGGIGVAGGLGGDD229 pKa = 4.35GGAGGAGGAGGANSQLLSLWGSAGAGGAGGIGGAGGAGTDD269 pKa = 4.12GSAGGIGADD278 pKa = 3.99GGAGGHH284 pKa = 6.27GGVGGAGGANNAWLGGAAGSGGTGGSGGTGGLGGAGGSGIGYY326 pKa = 7.74STGGGAGGAGGAGGDD341 pKa = 3.83AGTGGAAGATAFLGSAGVAGSAGNGGHH368 pKa = 6.67GGNGGSGADD377 pKa = 3.66GDD379 pKa = 3.97IGVAGSGAGADD390 pKa = 3.98GGNGFAGGTGGDD402 pKa = 3.91GGTGGNTTGGSNGSGGNAGHH422 pKa = 7.08GGNAGDD428 pKa = 4.06GAAGNTGVGFATKK441 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH477 pKa = 6.97GGNGGNGADD486 pKa = 4.09GDD488 pKa = 4.0IGVAGIGAGADD499 pKa = 3.56GGNGFAGGTGGDD511 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH531 pKa = 7.13GGNAGNGAAGNTGVGFATKK550 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH586 pKa = 6.97GGNGGNGADD595 pKa = 4.09GDD597 pKa = 4.0IGVAGIGAGADD608 pKa = 3.56GGNGFAGGTGGDD620 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH640 pKa = 7.13GGNAGNGAAGNTGVGFATKK659 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH695 pKa = 6.97GGNGGNGADD704 pKa = 4.09GDD706 pKa = 4.0IGVAGIGAGADD717 pKa = 3.56GGNGFAGGTGGDD729 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH749 pKa = 7.13GGNAGNGAAGNTGVGFATKK768 pKa = 10.2GGNGGNGGIGGNAGTAGLAGTGGTGGTTGAVGNAGHH804 pKa = 6.97GGNGGNGADD813 pKa = 4.09GDD815 pKa = 4.0IGVAGIGAGADD826 pKa = 3.56GGNGFAGGTGGDD838 pKa = 3.91GGTGGNTTGGTNGSGGNAGHH858 pKa = 7.13GGNAGNGADD867 pKa = 4.16GNVGDD872 pKa = 4.82YY873 pKa = 10.21YY874 pKa = 11.59VSGGTGGMGGDD885 pKa = 3.83GGDD888 pKa = 3.46AGTAGLAGTGGSGGTVGAYY907 pKa = 9.57SAGGAAGNGGTGGAGAQGAPAQFGSTGFAGGDD939 pKa = 3.31GGAGGDD945 pKa = 3.8GGTGGGGNGGTNGNGGHH962 pKa = 6.85GGTGGAGGQGGVGYY976 pKa = 9.23TDD978 pKa = 4.87DD979 pKa = 4.24GSNNATTGGDD989 pKa = 3.65GGDD992 pKa = 3.94GGQGGQGGVPGATGTGGTVGMAGNGGAGGAGGQGGDD1028 pKa = 3.43TTTSGNSAGFGGQGGQGGQGGAGLSGLTGGDD1059 pKa = 4.1GYY1061 pKa = 11.39DD1062 pKa = 3.8GGQGGAGGTGGAGGDD1077 pKa = 3.84AVSGAYY1083 pKa = 9.57NGHH1086 pKa = 6.59GGAGGDD1092 pKa = 4.02GGAGGQGGTGHH1103 pKa = 7.56ADD1105 pKa = 3.3DD1106 pKa = 5.27GSNNAGAGGNGGMGGQGGQGGGAGAGAGAGSGAGSAGTGGDD1147 pKa = 3.6GGTGGDD1153 pKa = 3.78GGATTTIGHH1162 pKa = 5.6TAGAGGVGGQGGDD1175 pKa = 3.76GGDD1178 pKa = 3.88GLADD1182 pKa = 3.64AASSGGKK1189 pKa = 9.99GGAGGLGGNGGDD1201 pKa = 3.83AVSGAYY1207 pKa = 9.75NGQGGMGGQGGTGGQGGDD1225 pKa = 4.05AGPEE1229 pKa = 4.09GNGGNGGNGGDD1240 pKa = 4.07GGTAGAAGTGGLGGTAGTAGAGGTGGGGGQGGDD1273 pKa = 3.27TTTAGHH1279 pKa = 5.39TAGWGGNGGYY1289 pKa = 10.11GGQGGGGAAGPAGGSGSDD1307 pKa = 3.77GGQGGQGGAGGTGGDD1322 pKa = 3.67AVAGATNGSGGTGGYY1337 pKa = 9.65GGQGGQGGAGGTDD1350 pKa = 3.2DD1351 pKa = 5.4GGDD1354 pKa = 3.41NAGTGGAGGTGGTGGAAGATGSGGTVDD1381 pKa = 4.38LAFPGTAGHH1390 pKa = 6.65GGDD1393 pKa = 4.05GGAGGGGGATTIVGHH1408 pKa = 5.14TAGTGGSGGQGGEE1421 pKa = 3.99GGAAADD1427 pKa = 3.82TGVPGAAGSTGGFGGTGGAGGAGGDD1452 pKa = 3.73AVAGATNGSGGAGGQGGAGAKK1473 pKa = 9.92GGQGYY1478 pKa = 10.67DD1479 pKa = 3.9DD1480 pKa = 5.81DD1481 pKa = 4.72GANNAGVGGNGGEE1494 pKa = 4.41GGTGGSGGAAGTGGDD1509 pKa = 3.95GGTAGAAGTGGTGGDD1524 pKa = 3.73GGKK1527 pKa = 10.64GGATTISGHH1536 pKa = 5.23HH1537 pKa = 6.56AGYY1540 pKa = 10.29GGNAGGGGQGADD1552 pKa = 3.71GAAGPAGGNGYY1563 pKa = 10.62DD1564 pKa = 4.03GGQGGAGGTGGQGGDD1579 pKa = 3.37AVAGATNGYY1588 pKa = 9.06GGQGGIGGTGGQGGGGGADD1607 pKa = 3.93DD1608 pKa = 5.2GSDD1611 pKa = 3.21NAGAGGSGGAGGLGGQSGAASTGGIVGTGGTGFAGDD1647 pKa = 4.43GGDD1650 pKa = 4.24GGAGGQGGATAVSGHH1665 pKa = 5.23TAGAGGTGGTGGTGGDD1681 pKa = 4.51GIDD1684 pKa = 4.33GPADD1688 pKa = 3.73RR1689 pKa = 11.84PGGRR1693 pKa = 11.84GGQGGHH1699 pKa = 6.5GGDD1702 pKa = 3.75GGSGGSAVAGATNGSGGAGGAGGTGGQGGNGYY1734 pKa = 10.56DD1735 pKa = 4.97DD1736 pKa = 5.59DD1737 pKa = 6.57GSDD1740 pKa = 3.29AAGAGGAGGDD1750 pKa = 3.72GGVGGTAGAVGSGGSGGTAGTAGTGGDD1777 pKa = 3.95GGDD1780 pKa = 3.74GGKK1783 pKa = 10.65GGATTTIGSSAGSGGDD1799 pKa = 3.41GGNGGLGGAGAIGPAGADD1817 pKa = 3.45GSTGGQGGAGGQGGDD1832 pKa = 3.98GGDD1835 pKa = 3.56AVAGATNGLGGNGGDD1850 pKa = 4.29GGSGGQGGAGGVDD1863 pKa = 3.6DD1864 pKa = 5.46GGGAGTGGQGGEE1876 pKa = 4.31GGQAGDD1882 pKa = 4.15GGVSGSGGLFGGGGDD1897 pKa = 4.27GGHH1900 pKa = 6.93GGQGGLGGTGGDD1912 pKa = 3.59SALGTGGDD1920 pKa = 4.15GGQGGQGGTGGTGGYY1935 pKa = 10.1GAFEE1939 pKa = 4.57GGFGGDD1945 pKa = 3.77GGDD1948 pKa = 3.79GGTGGTGGVGQSGLPGTGGTIGIAGVPGSLPSGSTGGAGGDD1989 pKa = 3.94GGDD1992 pKa = 4.58GGDD1995 pKa = 4.59GGPGSPP2001 pKa = 4.79
Molecular weight: 160.6 kDa
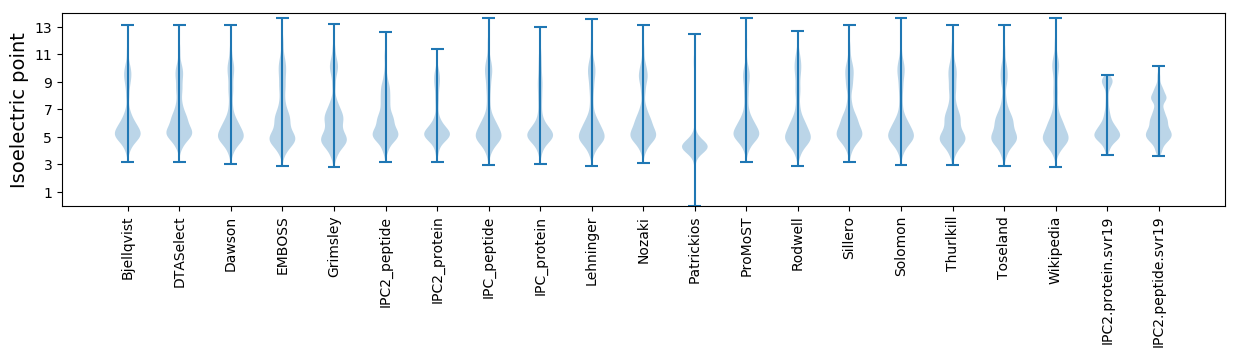
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2HNW0|B2HNW0_MYCMM PPE family protein PPE3 OS=Mycobacterium marinum (strain ATCC BAA-535 / M) OX=216594 GN=MMAR_0538 PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVTGRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.63GRR42 pKa = 11.84RR43 pKa = 11.84ALTAA47 pKa = 3.88
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVTGRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.63GRR42 pKa = 11.84RR43 pKa = 11.84ALTAA47 pKa = 3.88
Molecular weight: 5.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1984709 |
32 |
9858 |
366.3 |
38.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.223 ± 0.053 | 0.821 ± 0.012 |
5.812 ± 0.029 | 4.773 ± 0.036 |
3.017 ± 0.022 | 10.606 ± 0.176 |
2.228 ± 0.035 | 4.277 ± 0.024 |
1.941 ± 0.024 | 9.696 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.98 ± 0.017 | 2.585 ± 0.041 |
5.758 ± 0.046 | 3.235 ± 0.018 |
6.713 ± 0.051 | 5.665 ± 0.027 |
5.992 ± 0.056 | 8.127 ± 0.044 |
1.457 ± 0.014 | 2.095 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
