
Facklamia languida CCUG 37842
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Aerococcaceae; Facklamia; Facklamia languida
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
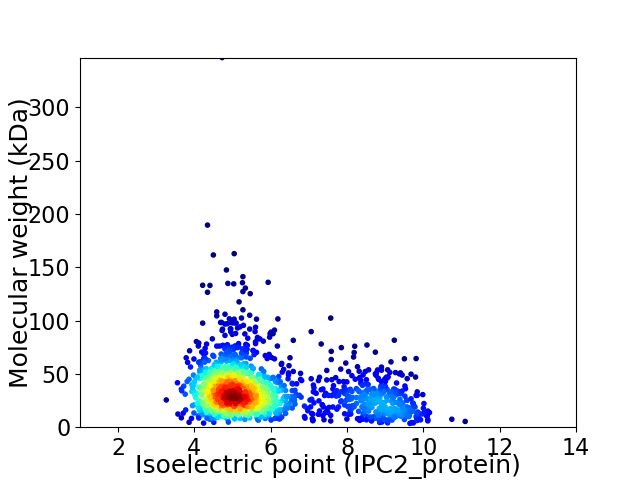
Virtual 2D-PAGE plot for 1527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H3NGR0|H3NGR0_9LACT SSD domain-containing protein OS=Facklamia languida CCUG 37842 OX=883113 GN=HMPREF9708_00049 PE=4 SV=1
MM1 pKa = 7.56NKK3 pKa = 7.52MTFLKK8 pKa = 10.58KK9 pKa = 9.61LTLAALFANFLCPPLTHH26 pKa = 6.52AQEE29 pKa = 4.48DD30 pKa = 4.11QATEE34 pKa = 4.07DD35 pKa = 3.37TSVTNEE41 pKa = 3.85EE42 pKa = 4.06GLDD45 pKa = 4.0LIEE48 pKa = 5.65SEE50 pKa = 5.35ASLEE54 pKa = 4.11DD55 pKa = 3.82LMDD58 pKa = 6.55DD59 pKa = 5.08IIQANQSVQSVVQEE73 pKa = 4.02IEE75 pKa = 4.05LDD77 pKa = 3.77SKK79 pKa = 11.22VSQEE83 pKa = 4.22GEE85 pKa = 3.53EE86 pKa = 4.48DD87 pKa = 3.78LFNQKK92 pKa = 10.1AVMEE96 pKa = 5.06AIYY99 pKa = 10.72GEE101 pKa = 4.25EE102 pKa = 4.24GQVDD106 pKa = 4.05GAYY109 pKa = 9.47IYY111 pKa = 11.06SLYY114 pKa = 10.28DD115 pKa = 3.2QNGEE119 pKa = 3.97QMIRR123 pKa = 11.84EE124 pKa = 3.98QLMPTNEE131 pKa = 3.7KK132 pKa = 9.85TIYY135 pKa = 9.71TRR137 pKa = 11.84FNEE140 pKa = 4.33EE141 pKa = 3.44EE142 pKa = 4.15WEE144 pKa = 4.12KK145 pKa = 11.53DD146 pKa = 3.02STEE149 pKa = 4.07YY150 pKa = 10.7TDD152 pKa = 3.15QDD154 pKa = 3.96QYY156 pKa = 12.01FLTPNYY162 pKa = 9.94HH163 pKa = 6.8HH164 pKa = 7.34LLADD168 pKa = 3.95LLEE171 pKa = 4.54LEE173 pKa = 4.78SYY175 pKa = 8.02YY176 pKa = 10.85TLYY179 pKa = 11.04EE180 pKa = 4.29DD181 pKa = 3.84ANSYY185 pKa = 10.72ILITKK190 pKa = 9.82DD191 pKa = 2.71QDD193 pKa = 3.02ADD195 pKa = 4.0LFHH198 pKa = 6.86VLQPHH203 pKa = 7.03FDD205 pKa = 3.66LQLTGMEE212 pKa = 4.61PEE214 pKa = 4.69AFHH217 pKa = 6.67QGVFLSINKK226 pKa = 7.97DD227 pKa = 3.15TLLFEE232 pKa = 5.79DD233 pKa = 4.67LMFVMTYY240 pKa = 11.1DD241 pKa = 4.42DD242 pKa = 4.69GSSSLEE248 pKa = 3.85MSAVAIMSEE257 pKa = 3.87WDD259 pKa = 3.41KK260 pKa = 11.79LNASDD265 pKa = 4.11YY266 pKa = 11.57QMDD269 pKa = 3.51QSEE272 pKa = 4.5PGQEE276 pKa = 4.29EE277 pKa = 3.89IDD279 pKa = 3.77QEE281 pKa = 4.39EE282 pKa = 4.37VDD284 pKa = 3.8QEE286 pKa = 4.16EE287 pKa = 4.56SSLEE291 pKa = 3.93EE292 pKa = 3.96TDD294 pKa = 4.63QEE296 pKa = 4.46EE297 pKa = 4.51ADD299 pKa = 3.63QEE301 pKa = 4.2ASQEE305 pKa = 4.22EE306 pKa = 4.78TSQEE310 pKa = 3.78
MM1 pKa = 7.56NKK3 pKa = 7.52MTFLKK8 pKa = 10.58KK9 pKa = 9.61LTLAALFANFLCPPLTHH26 pKa = 6.52AQEE29 pKa = 4.48DD30 pKa = 4.11QATEE34 pKa = 4.07DD35 pKa = 3.37TSVTNEE41 pKa = 3.85EE42 pKa = 4.06GLDD45 pKa = 4.0LIEE48 pKa = 5.65SEE50 pKa = 5.35ASLEE54 pKa = 4.11DD55 pKa = 3.82LMDD58 pKa = 6.55DD59 pKa = 5.08IIQANQSVQSVVQEE73 pKa = 4.02IEE75 pKa = 4.05LDD77 pKa = 3.77SKK79 pKa = 11.22VSQEE83 pKa = 4.22GEE85 pKa = 3.53EE86 pKa = 4.48DD87 pKa = 3.78LFNQKK92 pKa = 10.1AVMEE96 pKa = 5.06AIYY99 pKa = 10.72GEE101 pKa = 4.25EE102 pKa = 4.24GQVDD106 pKa = 4.05GAYY109 pKa = 9.47IYY111 pKa = 11.06SLYY114 pKa = 10.28DD115 pKa = 3.2QNGEE119 pKa = 3.97QMIRR123 pKa = 11.84EE124 pKa = 3.98QLMPTNEE131 pKa = 3.7KK132 pKa = 9.85TIYY135 pKa = 9.71TRR137 pKa = 11.84FNEE140 pKa = 4.33EE141 pKa = 3.44EE142 pKa = 4.15WEE144 pKa = 4.12KK145 pKa = 11.53DD146 pKa = 3.02STEE149 pKa = 4.07YY150 pKa = 10.7TDD152 pKa = 3.15QDD154 pKa = 3.96QYY156 pKa = 12.01FLTPNYY162 pKa = 9.94HH163 pKa = 6.8HH164 pKa = 7.34LLADD168 pKa = 3.95LLEE171 pKa = 4.54LEE173 pKa = 4.78SYY175 pKa = 8.02YY176 pKa = 10.85TLYY179 pKa = 11.04EE180 pKa = 4.29DD181 pKa = 3.84ANSYY185 pKa = 10.72ILITKK190 pKa = 9.82DD191 pKa = 2.71QDD193 pKa = 3.02ADD195 pKa = 4.0LFHH198 pKa = 6.86VLQPHH203 pKa = 7.03FDD205 pKa = 3.66LQLTGMEE212 pKa = 4.61PEE214 pKa = 4.69AFHH217 pKa = 6.67QGVFLSINKK226 pKa = 7.97DD227 pKa = 3.15TLLFEE232 pKa = 5.79DD233 pKa = 4.67LMFVMTYY240 pKa = 11.1DD241 pKa = 4.42DD242 pKa = 4.69GSSSLEE248 pKa = 3.85MSAVAIMSEE257 pKa = 3.87WDD259 pKa = 3.41KK260 pKa = 11.79LNASDD265 pKa = 4.11YY266 pKa = 11.57QMDD269 pKa = 3.51QSEE272 pKa = 4.5PGQEE276 pKa = 4.29EE277 pKa = 3.89IDD279 pKa = 3.77QEE281 pKa = 4.39EE282 pKa = 4.37VDD284 pKa = 3.8QEE286 pKa = 4.16EE287 pKa = 4.56SSLEE291 pKa = 3.93EE292 pKa = 3.96TDD294 pKa = 4.63QEE296 pKa = 4.46EE297 pKa = 4.51ADD299 pKa = 3.63QEE301 pKa = 4.2ASQEE305 pKa = 4.22EE306 pKa = 4.78TSQEE310 pKa = 3.78
Molecular weight: 35.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H3NIK5|H3NIK5_9LACT Alpha alpha-phosphotrehalase OS=Facklamia languida CCUG 37842 OX=883113 GN=HMPREF9708_00694 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.96RR4 pKa = 11.84TYY6 pKa = 9.59QPKK9 pKa = 9.46KK10 pKa = 7.41RR11 pKa = 11.84HH12 pKa = 5.45RR13 pKa = 11.84SKK15 pKa = 10.57VHH17 pKa = 5.58GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.41NGRR29 pKa = 11.84RR30 pKa = 11.84VLSKK34 pKa = 10.63RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.67GRR40 pKa = 11.84KK41 pKa = 8.83KK42 pKa = 10.59LSAA45 pKa = 3.95
MM1 pKa = 7.74SKK3 pKa = 8.96RR4 pKa = 11.84TYY6 pKa = 9.59QPKK9 pKa = 9.46KK10 pKa = 7.41RR11 pKa = 11.84HH12 pKa = 5.45RR13 pKa = 11.84SKK15 pKa = 10.57VHH17 pKa = 5.58GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.41NGRR29 pKa = 11.84RR30 pKa = 11.84VLSKK34 pKa = 10.63RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.67GRR40 pKa = 11.84KK41 pKa = 8.83KK42 pKa = 10.59LSAA45 pKa = 3.95
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
486735 |
31 |
3137 |
318.8 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.66 ± 0.07 | 0.599 ± 0.017 |
6.121 ± 0.048 | 6.872 ± 0.084 |
4.089 ± 0.049 | 6.45 ± 0.065 |
2.198 ± 0.035 | 7.211 ± 0.06 |
5.391 ± 0.058 | 10.342 ± 0.096 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.573 ± 0.026 | 3.95 ± 0.037 |
3.869 ± 0.042 | 5.698 ± 0.076 |
4.32 ± 0.051 | 5.823 ± 0.045 |
5.293 ± 0.072 | 6.782 ± 0.048 |
0.921 ± 0.023 | 3.837 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
