
Wenzhou tombus-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.81
Get precalculated fractions of proteins
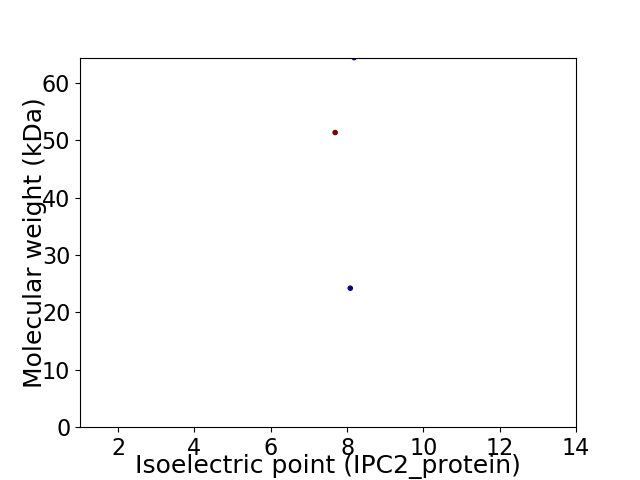
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH75|A0A1L3KH75_9VIRU Tombus_P33 domain-containing protein OS=Wenzhou tombus-like virus 5 OX=1923675 PE=4 SV=1
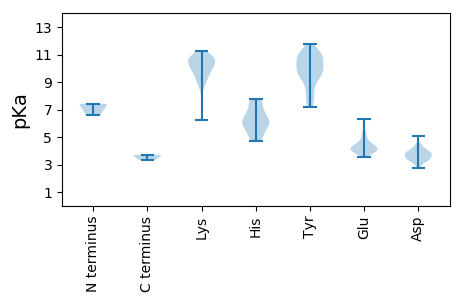
MM1 pKa = 7.42YY2 pKa = 9.45YY3 pKa = 10.43CKK5 pKa = 10.72VGDD8 pKa = 4.0TFLPAPEE15 pKa = 4.94PDD17 pKa = 3.69VNHH20 pKa = 5.61VFAGLRR26 pKa = 11.84EE27 pKa = 4.04FRR29 pKa = 11.84SKK31 pKa = 10.46LVHH34 pKa = 6.43RR35 pKa = 11.84MVSTPYY41 pKa = 9.93TIEE44 pKa = 4.14EE45 pKa = 4.11VVDD48 pKa = 3.72TYY50 pKa = 11.23KK51 pKa = 10.8GRR53 pKa = 11.84KK54 pKa = 6.26RR55 pKa = 11.84TIYY58 pKa = 9.56QQASEE63 pKa = 4.42SLNNTGLKK71 pKa = 9.74RR72 pKa = 11.84GHH74 pKa = 7.14AISDD78 pKa = 3.78CFVKK82 pKa = 10.71VEE84 pKa = 4.55KK85 pKa = 10.96GNTSKK90 pKa = 10.98APRR93 pKa = 11.84CIQPRR98 pKa = 11.84RR99 pKa = 11.84PEE101 pKa = 3.95YY102 pKa = 10.84NLVLGKK108 pKa = 9.93YY109 pKa = 8.75IKK111 pKa = 10.39KK112 pKa = 9.67IEE114 pKa = 3.69HH115 pKa = 5.29RR116 pKa = 11.84MYY118 pKa = 10.77RR119 pKa = 11.84RR120 pKa = 11.84IAKK123 pKa = 9.75IYY125 pKa = 9.84GDD127 pKa = 4.37GPTVMKK133 pKa = 10.49GYY135 pKa = 8.07TVQDD139 pKa = 3.23VARR142 pKa = 11.84IMRR145 pKa = 11.84GKK147 pKa = 9.03WDD149 pKa = 3.7SFSDD153 pKa = 4.03PVAVGLDD160 pKa = 3.27AVKK163 pKa = 10.78FDD165 pKa = 3.85MHH167 pKa = 7.39VSAAMLEE174 pKa = 4.47WEE176 pKa = 4.03HH177 pKa = 7.6SIYY180 pKa = 10.68KK181 pKa = 10.44AVTTDD186 pKa = 2.97TRR188 pKa = 11.84EE189 pKa = 4.07LNRR192 pKa = 11.84LLRR195 pKa = 11.84WQMFNKK201 pKa = 9.91GRR203 pKa = 11.84GRR205 pKa = 11.84CVDD208 pKa = 3.19GKK210 pKa = 9.48LTYY213 pKa = 8.37TVKK216 pKa = 10.67GKK218 pKa = 10.49RR219 pKa = 11.84FSGDD223 pKa = 2.78MNTALGNCIIMCGLVYY239 pKa = 10.1TYY241 pKa = 10.97LKK243 pKa = 9.93EE244 pKa = 4.27KK245 pKa = 10.24GMHH248 pKa = 6.21GKK250 pKa = 9.54LVNNGDD256 pKa = 3.76DD257 pKa = 3.26CVVFMEE263 pKa = 4.94RR264 pKa = 11.84GDD266 pKa = 3.72LEE268 pKa = 4.71GFVEE272 pKa = 6.34GLDD275 pKa = 3.2EE276 pKa = 4.08WFYY279 pKa = 11.51SYY281 pKa = 10.84GFRR284 pKa = 11.84MTSEE288 pKa = 4.04EE289 pKa = 4.08PVFDD293 pKa = 3.97FEE295 pKa = 6.25KK296 pKa = 10.69IEE298 pKa = 4.15FCQMRR303 pKa = 11.84PILGPNGWVMVRR315 pKa = 11.84NIKK318 pKa = 10.12AALVKK323 pKa = 10.71DD324 pKa = 4.1CLCTIDD330 pKa = 5.07LDD332 pKa = 4.37GVSARR337 pKa = 11.84KK338 pKa = 8.87WMYY341 pKa = 11.05AVGEE345 pKa = 4.42CGLALTSGIPIMQEE359 pKa = 3.89FYY361 pKa = 10.92LALMRR366 pKa = 11.84NGDD369 pKa = 3.66SSSNITQSLQFQSSGLRR386 pKa = 11.84HH387 pKa = 5.39MRR389 pKa = 11.84GEE391 pKa = 4.27LDD393 pKa = 3.34AKK395 pKa = 10.78ASAVSAEE402 pKa = 4.03ARR404 pKa = 11.84RR405 pKa = 11.84SVYY408 pKa = 9.55VAWGITPDD416 pKa = 3.47EE417 pKa = 4.05QVVLEE422 pKa = 3.96EE423 pKa = 4.93HH424 pKa = 5.66FRR426 pKa = 11.84SWVFDD431 pKa = 3.84PSPGDD436 pKa = 3.14IHH438 pKa = 7.74VSQPPIFHH446 pKa = 7.16GISLSWW452 pKa = 3.33
MM1 pKa = 7.42YY2 pKa = 9.45YY3 pKa = 10.43CKK5 pKa = 10.72VGDD8 pKa = 4.0TFLPAPEE15 pKa = 4.94PDD17 pKa = 3.69VNHH20 pKa = 5.61VFAGLRR26 pKa = 11.84EE27 pKa = 4.04FRR29 pKa = 11.84SKK31 pKa = 10.46LVHH34 pKa = 6.43RR35 pKa = 11.84MVSTPYY41 pKa = 9.93TIEE44 pKa = 4.14EE45 pKa = 4.11VVDD48 pKa = 3.72TYY50 pKa = 11.23KK51 pKa = 10.8GRR53 pKa = 11.84KK54 pKa = 6.26RR55 pKa = 11.84TIYY58 pKa = 9.56QQASEE63 pKa = 4.42SLNNTGLKK71 pKa = 9.74RR72 pKa = 11.84GHH74 pKa = 7.14AISDD78 pKa = 3.78CFVKK82 pKa = 10.71VEE84 pKa = 4.55KK85 pKa = 10.96GNTSKK90 pKa = 10.98APRR93 pKa = 11.84CIQPRR98 pKa = 11.84RR99 pKa = 11.84PEE101 pKa = 3.95YY102 pKa = 10.84NLVLGKK108 pKa = 9.93YY109 pKa = 8.75IKK111 pKa = 10.39KK112 pKa = 9.67IEE114 pKa = 3.69HH115 pKa = 5.29RR116 pKa = 11.84MYY118 pKa = 10.77RR119 pKa = 11.84RR120 pKa = 11.84IAKK123 pKa = 9.75IYY125 pKa = 9.84GDD127 pKa = 4.37GPTVMKK133 pKa = 10.49GYY135 pKa = 8.07TVQDD139 pKa = 3.23VARR142 pKa = 11.84IMRR145 pKa = 11.84GKK147 pKa = 9.03WDD149 pKa = 3.7SFSDD153 pKa = 4.03PVAVGLDD160 pKa = 3.27AVKK163 pKa = 10.78FDD165 pKa = 3.85MHH167 pKa = 7.39VSAAMLEE174 pKa = 4.47WEE176 pKa = 4.03HH177 pKa = 7.6SIYY180 pKa = 10.68KK181 pKa = 10.44AVTTDD186 pKa = 2.97TRR188 pKa = 11.84EE189 pKa = 4.07LNRR192 pKa = 11.84LLRR195 pKa = 11.84WQMFNKK201 pKa = 9.91GRR203 pKa = 11.84GRR205 pKa = 11.84CVDD208 pKa = 3.19GKK210 pKa = 9.48LTYY213 pKa = 8.37TVKK216 pKa = 10.67GKK218 pKa = 10.49RR219 pKa = 11.84FSGDD223 pKa = 2.78MNTALGNCIIMCGLVYY239 pKa = 10.1TYY241 pKa = 10.97LKK243 pKa = 9.93EE244 pKa = 4.27KK245 pKa = 10.24GMHH248 pKa = 6.21GKK250 pKa = 9.54LVNNGDD256 pKa = 3.76DD257 pKa = 3.26CVVFMEE263 pKa = 4.94RR264 pKa = 11.84GDD266 pKa = 3.72LEE268 pKa = 4.71GFVEE272 pKa = 6.34GLDD275 pKa = 3.2EE276 pKa = 4.08WFYY279 pKa = 11.51SYY281 pKa = 10.84GFRR284 pKa = 11.84MTSEE288 pKa = 4.04EE289 pKa = 4.08PVFDD293 pKa = 3.97FEE295 pKa = 6.25KK296 pKa = 10.69IEE298 pKa = 4.15FCQMRR303 pKa = 11.84PILGPNGWVMVRR315 pKa = 11.84NIKK318 pKa = 10.12AALVKK323 pKa = 10.71DD324 pKa = 4.1CLCTIDD330 pKa = 5.07LDD332 pKa = 4.37GVSARR337 pKa = 11.84KK338 pKa = 8.87WMYY341 pKa = 11.05AVGEE345 pKa = 4.42CGLALTSGIPIMQEE359 pKa = 3.89FYY361 pKa = 10.92LALMRR366 pKa = 11.84NGDD369 pKa = 3.66SSSNITQSLQFQSSGLRR386 pKa = 11.84HH387 pKa = 5.39MRR389 pKa = 11.84GEE391 pKa = 4.27LDD393 pKa = 3.34AKK395 pKa = 10.78ASAVSAEE402 pKa = 4.03ARR404 pKa = 11.84RR405 pKa = 11.84SVYY408 pKa = 9.55VAWGITPDD416 pKa = 3.47EE417 pKa = 4.05QVVLEE422 pKa = 3.96EE423 pKa = 4.93HH424 pKa = 5.66FRR426 pKa = 11.84SWVFDD431 pKa = 3.84PSPGDD436 pKa = 3.14IHH438 pKa = 7.74VSQPPIFHH446 pKa = 7.16GISLSWW452 pKa = 3.33
Molecular weight: 51.36 kDa
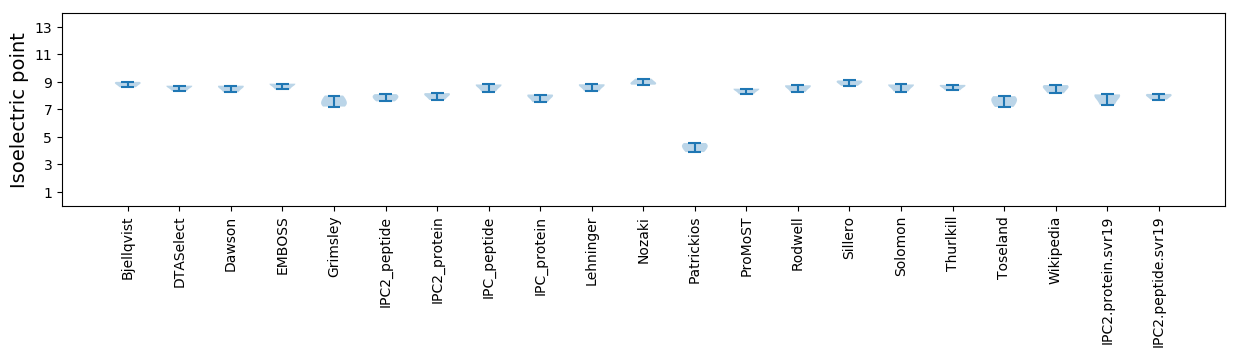
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGX3|A0A1L3KGX3_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 5 OX=1923675 PE=4 SV=1
MM1 pKa = 6.64GTAAVTSHH9 pKa = 6.33SLYY12 pKa = 10.79NSNLLGCATCAGSSTRR28 pKa = 11.84KK29 pKa = 9.23LAPLVQRR36 pKa = 11.84RR37 pKa = 11.84VGAFMWPGVSRR48 pKa = 11.84LMSRR52 pKa = 11.84LCWRR56 pKa = 11.84STSVAGCLTLPPVIFTYY73 pKa = 10.85HH74 pKa = 6.03NLPYY78 pKa = 10.51FMALAYY84 pKa = 9.82HH85 pKa = 6.72GNYY88 pKa = 9.29VGPGWSAGKK97 pKa = 9.35YY98 pKa = 7.32QSSVEE103 pKa = 4.09SDD105 pKa = 3.6VLPVDD110 pKa = 4.12EE111 pKa = 5.96FDD113 pKa = 3.77ATGKK117 pKa = 10.11RR118 pKa = 11.84HH119 pKa = 6.01DD120 pKa = 3.64AVYY123 pKa = 10.97ARR125 pKa = 11.84GGDD128 pKa = 3.37RR129 pKa = 11.84TRR131 pKa = 11.84ADD133 pKa = 3.48FEE135 pKa = 4.09FAYY138 pKa = 10.83DD139 pKa = 4.45NLTTLNPKK147 pKa = 9.78RR148 pKa = 11.84WLAGALVGAQGLCRR162 pKa = 11.84FATGLNGITEE172 pKa = 4.35EE173 pKa = 4.62AEE175 pKa = 4.26GSQRR179 pKa = 11.84GVTKK183 pKa = 10.64HH184 pKa = 5.96SSQGLDD190 pKa = 2.85KK191 pKa = 10.7DD192 pKa = 4.22YY193 pKa = 11.26PPLSIDD199 pKa = 3.17NMKK202 pKa = 10.66NKK204 pKa = 9.97RR205 pKa = 11.84NRR207 pKa = 11.84IFAGDD212 pKa = 3.76SSHH215 pKa = 7.75LVMDD219 pKa = 4.06SNGRR223 pKa = 11.84RR224 pKa = 11.84EE225 pKa = 3.91VEE227 pKa = 4.6RR228 pKa = 11.84IDD230 pKa = 3.59KK231 pKa = 10.66QITKK235 pKa = 9.9SYY237 pKa = 10.45QKK239 pKa = 10.41RR240 pKa = 11.84KK241 pKa = 8.7PLKK244 pKa = 10.14QRR246 pKa = 11.84VQQAFSPLKK255 pKa = 7.71TTAAPVSIGTTLVAVPPQTVKK276 pKa = 10.06TGNGVCIAGRR286 pKa = 11.84EE287 pKa = 4.5FITQVYY293 pKa = 7.28GTNNDD298 pKa = 3.39SFQVAGLAPVHH309 pKa = 6.13PAYY312 pKa = 10.86YY313 pKa = 9.45PASTMGNISRR323 pKa = 11.84SYY325 pKa = 9.06QYY327 pKa = 11.13YY328 pKa = 9.13RR329 pKa = 11.84FRR331 pKa = 11.84KK332 pKa = 8.73IVAHH336 pKa = 6.25FVTRR340 pKa = 11.84QPTSVTGEE348 pKa = 3.53IVMCYY353 pKa = 9.79SKK355 pKa = 11.2NILDD359 pKa = 4.02PPEE362 pKa = 5.33NGDD365 pKa = 3.5SGSFLPRR372 pKa = 11.84AMTRR376 pKa = 11.84GNAVIGPLWQNHH388 pKa = 4.7SVEE391 pKa = 4.35IPLDD395 pKa = 3.42GTFRR399 pKa = 11.84LVDD402 pKa = 3.62AFNAGTFADD411 pKa = 3.9NVNGEE416 pKa = 4.4VQVYY420 pKa = 7.21TQTNVDD426 pKa = 3.9DD427 pKa = 3.93TCGYY431 pKa = 10.05LVWDD435 pKa = 3.89YY436 pKa = 11.77VLEE439 pKa = 4.35FSNTMFTPHH448 pKa = 6.09STSLPLVSGPGTKK461 pKa = 8.94YY462 pKa = 8.7TVSVAASAANDD473 pKa = 3.84ALIGTSSLITAYY485 pKa = 9.82PNQTVWSLALDD496 pKa = 4.26LSASSAGTGATLANLVNVFANYY518 pKa = 7.62HH519 pKa = 4.68TTTTALSQAGTNLPLVDD536 pKa = 3.92GQRR539 pKa = 11.84IFAMVVGGVLYY550 pKa = 9.8FYY552 pKa = 10.97SSYY555 pKa = 9.73EE556 pKa = 3.78AAVAGMGSAQLYY568 pKa = 9.81IRR570 pKa = 11.84TATTTASTLIFNGYY584 pKa = 7.97LVRR587 pKa = 11.84LGPVEE592 pKa = 3.92LTAANN597 pKa = 3.62
MM1 pKa = 6.64GTAAVTSHH9 pKa = 6.33SLYY12 pKa = 10.79NSNLLGCATCAGSSTRR28 pKa = 11.84KK29 pKa = 9.23LAPLVQRR36 pKa = 11.84RR37 pKa = 11.84VGAFMWPGVSRR48 pKa = 11.84LMSRR52 pKa = 11.84LCWRR56 pKa = 11.84STSVAGCLTLPPVIFTYY73 pKa = 10.85HH74 pKa = 6.03NLPYY78 pKa = 10.51FMALAYY84 pKa = 9.82HH85 pKa = 6.72GNYY88 pKa = 9.29VGPGWSAGKK97 pKa = 9.35YY98 pKa = 7.32QSSVEE103 pKa = 4.09SDD105 pKa = 3.6VLPVDD110 pKa = 4.12EE111 pKa = 5.96FDD113 pKa = 3.77ATGKK117 pKa = 10.11RR118 pKa = 11.84HH119 pKa = 6.01DD120 pKa = 3.64AVYY123 pKa = 10.97ARR125 pKa = 11.84GGDD128 pKa = 3.37RR129 pKa = 11.84TRR131 pKa = 11.84ADD133 pKa = 3.48FEE135 pKa = 4.09FAYY138 pKa = 10.83DD139 pKa = 4.45NLTTLNPKK147 pKa = 9.78RR148 pKa = 11.84WLAGALVGAQGLCRR162 pKa = 11.84FATGLNGITEE172 pKa = 4.35EE173 pKa = 4.62AEE175 pKa = 4.26GSQRR179 pKa = 11.84GVTKK183 pKa = 10.64HH184 pKa = 5.96SSQGLDD190 pKa = 2.85KK191 pKa = 10.7DD192 pKa = 4.22YY193 pKa = 11.26PPLSIDD199 pKa = 3.17NMKK202 pKa = 10.66NKK204 pKa = 9.97RR205 pKa = 11.84NRR207 pKa = 11.84IFAGDD212 pKa = 3.76SSHH215 pKa = 7.75LVMDD219 pKa = 4.06SNGRR223 pKa = 11.84RR224 pKa = 11.84EE225 pKa = 3.91VEE227 pKa = 4.6RR228 pKa = 11.84IDD230 pKa = 3.59KK231 pKa = 10.66QITKK235 pKa = 9.9SYY237 pKa = 10.45QKK239 pKa = 10.41RR240 pKa = 11.84KK241 pKa = 8.7PLKK244 pKa = 10.14QRR246 pKa = 11.84VQQAFSPLKK255 pKa = 7.71TTAAPVSIGTTLVAVPPQTVKK276 pKa = 10.06TGNGVCIAGRR286 pKa = 11.84EE287 pKa = 4.5FITQVYY293 pKa = 7.28GTNNDD298 pKa = 3.39SFQVAGLAPVHH309 pKa = 6.13PAYY312 pKa = 10.86YY313 pKa = 9.45PASTMGNISRR323 pKa = 11.84SYY325 pKa = 9.06QYY327 pKa = 11.13YY328 pKa = 9.13RR329 pKa = 11.84FRR331 pKa = 11.84KK332 pKa = 8.73IVAHH336 pKa = 6.25FVTRR340 pKa = 11.84QPTSVTGEE348 pKa = 3.53IVMCYY353 pKa = 9.79SKK355 pKa = 11.2NILDD359 pKa = 4.02PPEE362 pKa = 5.33NGDD365 pKa = 3.5SGSFLPRR372 pKa = 11.84AMTRR376 pKa = 11.84GNAVIGPLWQNHH388 pKa = 4.7SVEE391 pKa = 4.35IPLDD395 pKa = 3.42GTFRR399 pKa = 11.84LVDD402 pKa = 3.62AFNAGTFADD411 pKa = 3.9NVNGEE416 pKa = 4.4VQVYY420 pKa = 7.21TQTNVDD426 pKa = 3.9DD427 pKa = 3.93TCGYY431 pKa = 10.05LVWDD435 pKa = 3.89YY436 pKa = 11.77VLEE439 pKa = 4.35FSNTMFTPHH448 pKa = 6.09STSLPLVSGPGTKK461 pKa = 8.94YY462 pKa = 8.7TVSVAASAANDD473 pKa = 3.84ALIGTSSLITAYY485 pKa = 9.82PNQTVWSLALDD496 pKa = 4.26LSASSAGTGATLANLVNVFANYY518 pKa = 7.62HH519 pKa = 4.68TTTTALSQAGTNLPLVDD536 pKa = 3.92GQRR539 pKa = 11.84IFAMVVGGVLYY550 pKa = 9.8FYY552 pKa = 10.97SSYY555 pKa = 9.73EE556 pKa = 3.78AAVAGMGSAQLYY568 pKa = 9.81IRR570 pKa = 11.84TATTTASTLIFNGYY584 pKa = 7.97LVRR587 pKa = 11.84LGPVEE592 pKa = 3.92LTAANN597 pKa = 3.62
Molecular weight: 64.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1266 |
217 |
597 |
422.0 |
46.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.847 ± 1.456 | 2.054 ± 0.476 |
4.581 ± 0.509 | 4.581 ± 1.211 |
3.949 ± 0.223 | 7.978 ± 0.775 |
1.975 ± 0.204 | 3.87 ± 0.52 |
4.423 ± 0.915 | 7.741 ± 0.398 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.923 ± 0.617 | 5.134 ± 0.905 |
4.423 ± 0.383 | 3.397 ± 0.318 |
6.319 ± 0.763 | 6.714 ± 1.122 |
6.556 ± 1.577 | 8.847 ± 0.575 |
1.501 ± 0.229 | 4.186 ± 0.418 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
