
Canid alphaherpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Varicellovirus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
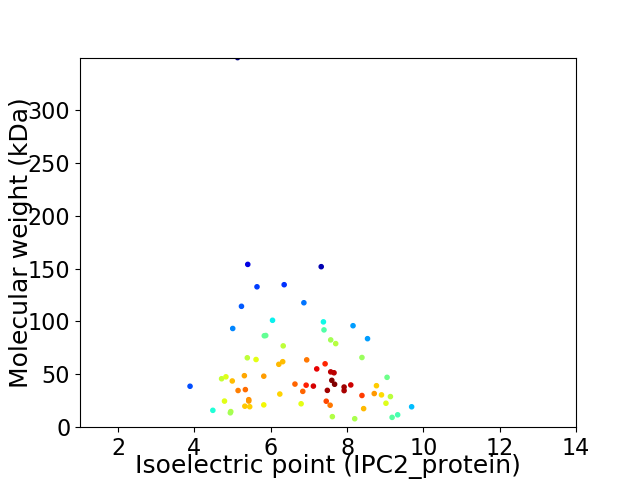
Virtual 2D-PAGE plot for 72 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A172DSI5|A0A172DSI5_9ALPH Membrane protein UL56 OS=Canid alphaherpesvirus 1 OX=170325 GN=UL56 PE=4 SV=1
MM1 pKa = 7.76ADD3 pKa = 3.35EE4 pKa = 5.43EE5 pKa = 5.12YY6 pKa = 11.19NCTICLEE13 pKa = 4.3PPKK16 pKa = 11.06NMTVTMSCLHH26 pKa = 6.23KK27 pKa = 10.53FCYY30 pKa = 10.1DD31 pKa = 4.22CLSEE35 pKa = 4.01WTKK38 pKa = 11.0VSNTCPLCKK47 pKa = 10.29SIIQSMIHH55 pKa = 6.65SINDD59 pKa = 3.27DD60 pKa = 3.8KK61 pKa = 11.35EE62 pKa = 4.13FKK64 pKa = 9.96EE65 pKa = 3.87IKK67 pKa = 9.78IVSEE71 pKa = 4.36SIEE74 pKa = 4.48DD75 pKa = 3.78STDD78 pKa = 3.38LLMEE82 pKa = 4.48EE83 pKa = 3.88NAQRR87 pKa = 11.84FFNSDD92 pKa = 3.33EE93 pKa = 3.97EE94 pKa = 4.49DD95 pKa = 3.99ANDD98 pKa = 5.24DD99 pKa = 4.26DD100 pKa = 6.47DD101 pKa = 5.24RR102 pKa = 11.84PLWGEE107 pKa = 4.71DD108 pKa = 3.17YY109 pKa = 11.31DD110 pKa = 4.38EE111 pKa = 5.37NYY113 pKa = 10.6SIYY116 pKa = 10.18QSEE119 pKa = 4.29PASSQIQPPSHH130 pKa = 6.68NSMEE134 pKa = 4.25QILFRR139 pKa = 11.84NPNTRR144 pKa = 11.84EE145 pKa = 3.54LLIRR149 pKa = 11.84FSYY152 pKa = 9.4NRR154 pKa = 11.84LMVYY158 pKa = 10.06YY159 pKa = 9.0EE160 pKa = 4.24HH161 pKa = 7.46EE162 pKa = 4.62GNAEE166 pKa = 4.1DD167 pKa = 3.48VTEE170 pKa = 5.17IIMEE174 pKa = 5.0FIDD177 pKa = 4.99EE178 pKa = 4.37YY179 pKa = 11.65GLDD182 pKa = 3.67RR183 pKa = 11.84DD184 pKa = 4.19EE185 pKa = 4.32LTEE188 pKa = 4.05LLEE191 pKa = 4.73PLLQTYY197 pKa = 7.45TIRR200 pKa = 11.84FVNSFFTRR208 pKa = 11.84INLLSRR214 pKa = 11.84RR215 pKa = 11.84PFRR218 pKa = 11.84ISTGVQFLDD227 pKa = 5.54DD228 pKa = 6.05DD229 pKa = 6.18DD230 pKa = 7.46DD231 pKa = 7.11DD232 pKa = 7.03DD233 pKa = 6.47DD234 pKa = 4.84EE235 pKa = 6.14NSVDD239 pKa = 4.04SEE241 pKa = 4.49SDD243 pKa = 3.41SSSSLCTDD251 pKa = 4.4DD252 pKa = 5.5LTIPDD257 pKa = 4.67DD258 pKa = 4.01TQSDD262 pKa = 3.5ISDD265 pKa = 3.82LDD267 pKa = 3.49EE268 pKa = 4.5TNISTGQSVRR278 pKa = 11.84EE279 pKa = 4.02NYY281 pKa = 7.67RR282 pKa = 11.84TRR284 pKa = 11.84LRR286 pKa = 11.84SYY288 pKa = 9.4TEE290 pKa = 3.15NSTRR294 pKa = 11.84YY295 pKa = 10.05SRR297 pKa = 11.84VLGNIMSPTGGSYY310 pKa = 10.21EE311 pKa = 4.05PEE313 pKa = 3.47ISEE316 pKa = 3.96EE317 pKa = 4.21PEE319 pKa = 3.63NVEE322 pKa = 5.61IIDD325 pKa = 3.95LTIDD329 pKa = 3.29SDD331 pKa = 4.63DD332 pKa = 5.4DD333 pKa = 3.64IGG335 pKa = 4.75
MM1 pKa = 7.76ADD3 pKa = 3.35EE4 pKa = 5.43EE5 pKa = 5.12YY6 pKa = 11.19NCTICLEE13 pKa = 4.3PPKK16 pKa = 11.06NMTVTMSCLHH26 pKa = 6.23KK27 pKa = 10.53FCYY30 pKa = 10.1DD31 pKa = 4.22CLSEE35 pKa = 4.01WTKK38 pKa = 11.0VSNTCPLCKK47 pKa = 10.29SIIQSMIHH55 pKa = 6.65SINDD59 pKa = 3.27DD60 pKa = 3.8KK61 pKa = 11.35EE62 pKa = 4.13FKK64 pKa = 9.96EE65 pKa = 3.87IKK67 pKa = 9.78IVSEE71 pKa = 4.36SIEE74 pKa = 4.48DD75 pKa = 3.78STDD78 pKa = 3.38LLMEE82 pKa = 4.48EE83 pKa = 3.88NAQRR87 pKa = 11.84FFNSDD92 pKa = 3.33EE93 pKa = 3.97EE94 pKa = 4.49DD95 pKa = 3.99ANDD98 pKa = 5.24DD99 pKa = 4.26DD100 pKa = 6.47DD101 pKa = 5.24RR102 pKa = 11.84PLWGEE107 pKa = 4.71DD108 pKa = 3.17YY109 pKa = 11.31DD110 pKa = 4.38EE111 pKa = 5.37NYY113 pKa = 10.6SIYY116 pKa = 10.18QSEE119 pKa = 4.29PASSQIQPPSHH130 pKa = 6.68NSMEE134 pKa = 4.25QILFRR139 pKa = 11.84NPNTRR144 pKa = 11.84EE145 pKa = 3.54LLIRR149 pKa = 11.84FSYY152 pKa = 9.4NRR154 pKa = 11.84LMVYY158 pKa = 10.06YY159 pKa = 9.0EE160 pKa = 4.24HH161 pKa = 7.46EE162 pKa = 4.62GNAEE166 pKa = 4.1DD167 pKa = 3.48VTEE170 pKa = 5.17IIMEE174 pKa = 5.0FIDD177 pKa = 4.99EE178 pKa = 4.37YY179 pKa = 11.65GLDD182 pKa = 3.67RR183 pKa = 11.84DD184 pKa = 4.19EE185 pKa = 4.32LTEE188 pKa = 4.05LLEE191 pKa = 4.73PLLQTYY197 pKa = 7.45TIRR200 pKa = 11.84FVNSFFTRR208 pKa = 11.84INLLSRR214 pKa = 11.84RR215 pKa = 11.84PFRR218 pKa = 11.84ISTGVQFLDD227 pKa = 5.54DD228 pKa = 6.05DD229 pKa = 6.18DD230 pKa = 7.46DD231 pKa = 7.11DD232 pKa = 7.03DD233 pKa = 6.47DD234 pKa = 4.84EE235 pKa = 6.14NSVDD239 pKa = 4.04SEE241 pKa = 4.49SDD243 pKa = 3.41SSSSLCTDD251 pKa = 4.4DD252 pKa = 5.5LTIPDD257 pKa = 4.67DD258 pKa = 4.01TQSDD262 pKa = 3.5ISDD265 pKa = 3.82LDD267 pKa = 3.49EE268 pKa = 4.5TNISTGQSVRR278 pKa = 11.84EE279 pKa = 4.02NYY281 pKa = 7.67RR282 pKa = 11.84TRR284 pKa = 11.84LRR286 pKa = 11.84SYY288 pKa = 9.4TEE290 pKa = 3.15NSTRR294 pKa = 11.84YY295 pKa = 10.05SRR297 pKa = 11.84VLGNIMSPTGGSYY310 pKa = 10.21EE311 pKa = 4.05PEE313 pKa = 3.47ISEE316 pKa = 3.96EE317 pKa = 4.21PEE319 pKa = 3.63NVEE322 pKa = 5.61IIDD325 pKa = 3.95LTIDD329 pKa = 3.29SDD331 pKa = 4.63DD332 pKa = 5.4DD333 pKa = 3.64IGG335 pKa = 4.75
Molecular weight: 38.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A172DSI1|A0A172DSI1_9ALPH Virion host shutoff protein OS=Canid alphaherpesvirus 1 OX=170325 GN=UL41 PE=3 SV=1
MM1 pKa = 7.82ASFNPADD8 pKa = 4.44LKK10 pKa = 10.57TITAANLRR18 pKa = 11.84GLLPAQIITIINQSNHH34 pKa = 6.29PNNASPAEE42 pKa = 3.94ILDD45 pKa = 3.57AQRR48 pKa = 11.84NLLIGTSLSMVALRR62 pKa = 11.84QRR64 pKa = 11.84HH65 pKa = 5.51ANHH68 pKa = 6.92TIARR72 pKa = 11.84VPMFAEE78 pKa = 3.9YY79 pKa = 10.6DD80 pKa = 3.68GLFWARR86 pKa = 11.84PTIGLKK92 pKa = 9.08RR93 pKa = 11.84TFSPRR98 pKa = 11.84LTQIVPDD105 pKa = 3.87EE106 pKa = 4.12
MM1 pKa = 7.82ASFNPADD8 pKa = 4.44LKK10 pKa = 10.57TITAANLRR18 pKa = 11.84GLLPAQIITIINQSNHH34 pKa = 6.29PNNASPAEE42 pKa = 3.94ILDD45 pKa = 3.57AQRR48 pKa = 11.84NLLIGTSLSMVALRR62 pKa = 11.84QRR64 pKa = 11.84HH65 pKa = 5.51ANHH68 pKa = 6.92TIARR72 pKa = 11.84VPMFAEE78 pKa = 3.9YY79 pKa = 10.6DD80 pKa = 3.68GLFWARR86 pKa = 11.84PTIGLKK92 pKa = 9.08RR93 pKa = 11.84TFSPRR98 pKa = 11.84LTQIVPDD105 pKa = 3.87EE106 pKa = 4.12
Molecular weight: 11.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
35638 |
70 |
3080 |
495.0 |
56.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.31 ± 0.176 | 1.858 ± 0.145 |
5.211 ± 0.185 | 5.794 ± 0.213 |
4.978 ± 0.191 | 4.332 ± 0.178 |
1.844 ± 0.084 | 8.623 ± 0.258 |
6.378 ± 0.262 | 9.661 ± 0.215 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.032 ± 0.079 | 7.203 ± 0.2 |
4.737 ± 0.2 | 3.275 ± 0.136 |
4.265 ± 0.169 | 9.568 ± 0.273 |
6.075 ± 0.105 | 5.138 ± 0.15 |
0.901 ± 0.056 | 3.819 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
