
Parasphingorhabdus marina DSM 22363
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Parasphingorhabdus; Parasphingorhabdus marina
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
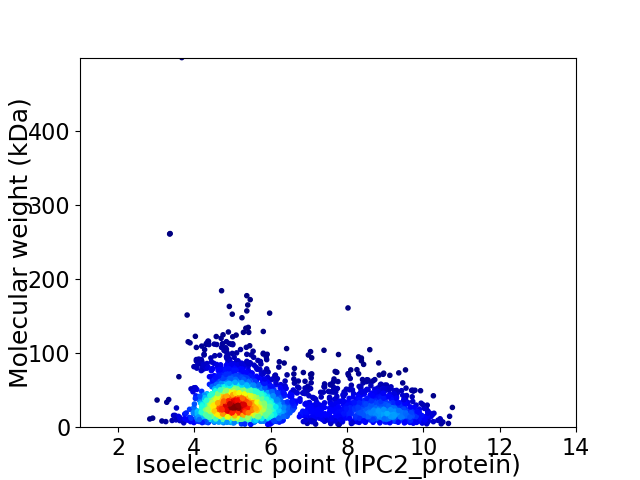
Virtual 2D-PAGE plot for 3398 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N6HFU3|A0A1N6HFU3_9SPHN Winged helix DNA-binding domain-containing protein OS=Parasphingorhabdus marina DSM 22363 OX=1123272 GN=SAMN02745824_3270 PE=4 SV=1
MM1 pKa = 7.49FVGTCDD7 pKa = 3.86SGWTRR12 pKa = 11.84SGAQSCVRR20 pKa = 11.84YY21 pKa = 9.04LAQLLLGLITLALFPAIAQAQTTTQYY47 pKa = 10.8TITDD51 pKa = 3.97PGVADD56 pKa = 6.11DD57 pKa = 3.96ITGSQICSSPLVQNFSVPDD76 pKa = 3.67NFTVSDD82 pKa = 3.57VDD84 pKa = 3.22IGVIATHH91 pKa = 5.68SWRR94 pKa = 11.84GDD96 pKa = 3.1MRR98 pKa = 11.84ITLEE102 pKa = 4.25SPLGTRR108 pKa = 11.84VRR110 pKa = 11.84VVNGSGGDD118 pKa = 3.28SGDD121 pKa = 3.75NFDD124 pKa = 5.18VRR126 pKa = 11.84LNDD129 pKa = 4.36GGTQLVNTDD138 pKa = 3.85PATGNHH144 pKa = 5.04NTGLPLYY151 pKa = 9.22EE152 pKa = 4.66NNFIPDD158 pKa = 3.46NALSAFNGEE167 pKa = 4.16NSTGTWRR174 pKa = 11.84LEE176 pKa = 3.61ICDD179 pKa = 4.27IFPSEE184 pKa = 4.8DD185 pKa = 3.19DD186 pKa = 4.2GNFLRR191 pKa = 11.84ADD193 pKa = 4.22LFLTDD198 pKa = 4.42TPPFADD204 pKa = 3.69LSVNKK209 pKa = 8.88TVNDD213 pKa = 3.59SSPNNGNNVTYY224 pKa = 10.06TITVTNASSSPSTATGIVVNDD245 pKa = 3.61QLPLGVTYY253 pKa = 10.97VSDD256 pKa = 3.84TGGGAYY262 pKa = 10.4NSGTGNWSVGTLAPGGSATLNIVANVTATSGATVTNAAEE301 pKa = 4.37VTVSSIFDD309 pKa = 3.92LDD311 pKa = 3.74STPGNGATGEE321 pKa = 4.3DD322 pKa = 4.73DD323 pKa = 4.15YY324 pKa = 11.88DD325 pKa = 3.94DD326 pKa = 5.43ASFTVAGTRR335 pKa = 11.84IAGTPPSLSTICSPANQILFDD356 pKa = 4.13WNGKK360 pKa = 8.76AWTTGSTNNNFTVAGIGVTNYY381 pKa = 10.99AITTDD386 pKa = 3.51VAFVTGSPAINSTNTGGQGATEE408 pKa = 3.76QSLFLNMNNNTRR420 pKa = 11.84SDD422 pKa = 3.26TSTTVLTLPTAVPGLQFTLFDD443 pKa = 3.54VDD445 pKa = 5.17FGAGQWADD453 pKa = 3.21KK454 pKa = 10.0VTVTGTFNGTTVIPTLTNGTANYY477 pKa = 10.19VIGNTAIGDD486 pKa = 3.94VGSDD490 pKa = 3.84GPDD493 pKa = 3.11PDD495 pKa = 4.17GNVVVTFLAPVDD507 pKa = 3.96TVTIVYY513 pKa = 9.97GNHH516 pKa = 4.11TTAPVNPGNQFMNIHH531 pKa = 6.85DD532 pKa = 4.44MNYY535 pKa = 9.29CQPDD539 pKa = 3.55TTFSVTKK546 pKa = 9.19ISSVISDD553 pKa = 3.74PVSIVDD559 pKa = 3.55GTSIPKK565 pKa = 9.9SIPGAIVQYY574 pKa = 10.52CILISNSGSATATDD588 pKa = 4.33IVATDD593 pKa = 3.81NLVGNYY599 pKa = 7.72TYY601 pKa = 10.29TAGSMQSGTNCNAAGTPEE619 pKa = 5.55DD620 pKa = 5.23DD621 pKa = 4.55DD622 pKa = 6.54ASDD625 pKa = 3.77GAEE628 pKa = 4.01SDD630 pKa = 3.92GVTASVIGTVLTANIASLGPSASTALTFRR659 pKa = 11.84VTVDD663 pKa = 2.78
MM1 pKa = 7.49FVGTCDD7 pKa = 3.86SGWTRR12 pKa = 11.84SGAQSCVRR20 pKa = 11.84YY21 pKa = 9.04LAQLLLGLITLALFPAIAQAQTTTQYY47 pKa = 10.8TITDD51 pKa = 3.97PGVADD56 pKa = 6.11DD57 pKa = 3.96ITGSQICSSPLVQNFSVPDD76 pKa = 3.67NFTVSDD82 pKa = 3.57VDD84 pKa = 3.22IGVIATHH91 pKa = 5.68SWRR94 pKa = 11.84GDD96 pKa = 3.1MRR98 pKa = 11.84ITLEE102 pKa = 4.25SPLGTRR108 pKa = 11.84VRR110 pKa = 11.84VVNGSGGDD118 pKa = 3.28SGDD121 pKa = 3.75NFDD124 pKa = 5.18VRR126 pKa = 11.84LNDD129 pKa = 4.36GGTQLVNTDD138 pKa = 3.85PATGNHH144 pKa = 5.04NTGLPLYY151 pKa = 9.22EE152 pKa = 4.66NNFIPDD158 pKa = 3.46NALSAFNGEE167 pKa = 4.16NSTGTWRR174 pKa = 11.84LEE176 pKa = 3.61ICDD179 pKa = 4.27IFPSEE184 pKa = 4.8DD185 pKa = 3.19DD186 pKa = 4.2GNFLRR191 pKa = 11.84ADD193 pKa = 4.22LFLTDD198 pKa = 4.42TPPFADD204 pKa = 3.69LSVNKK209 pKa = 8.88TVNDD213 pKa = 3.59SSPNNGNNVTYY224 pKa = 10.06TITVTNASSSPSTATGIVVNDD245 pKa = 3.61QLPLGVTYY253 pKa = 10.97VSDD256 pKa = 3.84TGGGAYY262 pKa = 10.4NSGTGNWSVGTLAPGGSATLNIVANVTATSGATVTNAAEE301 pKa = 4.37VTVSSIFDD309 pKa = 3.92LDD311 pKa = 3.74STPGNGATGEE321 pKa = 4.3DD322 pKa = 4.73DD323 pKa = 4.15YY324 pKa = 11.88DD325 pKa = 3.94DD326 pKa = 5.43ASFTVAGTRR335 pKa = 11.84IAGTPPSLSTICSPANQILFDD356 pKa = 4.13WNGKK360 pKa = 8.76AWTTGSTNNNFTVAGIGVTNYY381 pKa = 10.99AITTDD386 pKa = 3.51VAFVTGSPAINSTNTGGQGATEE408 pKa = 3.76QSLFLNMNNNTRR420 pKa = 11.84SDD422 pKa = 3.26TSTTVLTLPTAVPGLQFTLFDD443 pKa = 3.54VDD445 pKa = 5.17FGAGQWADD453 pKa = 3.21KK454 pKa = 10.0VTVTGTFNGTTVIPTLTNGTANYY477 pKa = 10.19VIGNTAIGDD486 pKa = 3.94VGSDD490 pKa = 3.84GPDD493 pKa = 3.11PDD495 pKa = 4.17GNVVVTFLAPVDD507 pKa = 3.96TVTIVYY513 pKa = 9.97GNHH516 pKa = 4.11TTAPVNPGNQFMNIHH531 pKa = 6.85DD532 pKa = 4.44MNYY535 pKa = 9.29CQPDD539 pKa = 3.55TTFSVTKK546 pKa = 9.19ISSVISDD553 pKa = 3.74PVSIVDD559 pKa = 3.55GTSIPKK565 pKa = 9.9SIPGAIVQYY574 pKa = 10.52CILISNSGSATATDD588 pKa = 4.33IVATDD593 pKa = 3.81NLVGNYY599 pKa = 7.72TYY601 pKa = 10.29TAGSMQSGTNCNAAGTPEE619 pKa = 5.55DD620 pKa = 5.23DD621 pKa = 4.55DD622 pKa = 6.54ASDD625 pKa = 3.77GAEE628 pKa = 4.01SDD630 pKa = 3.92GVTASVIGTVLTANIASLGPSASTALTFRR659 pKa = 11.84VTVDD663 pKa = 2.78
Molecular weight: 68.18 kDa
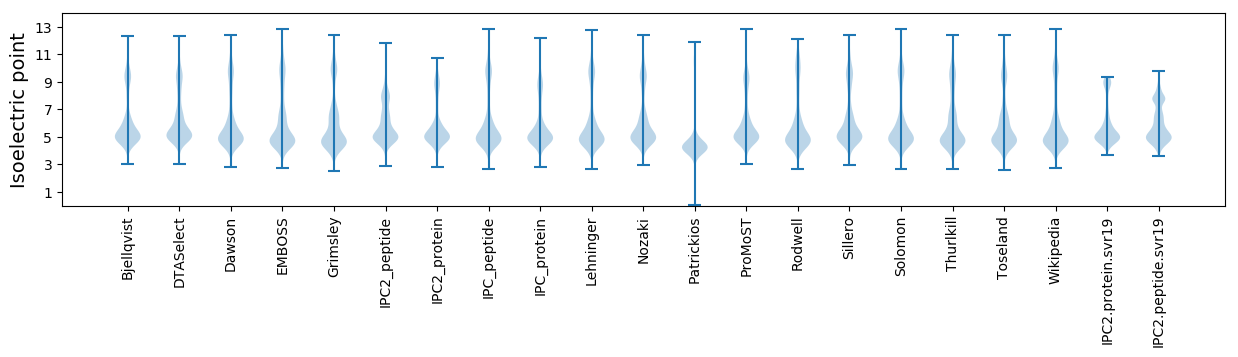
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N6CMI9|A0A1N6CMI9_9SPHN Iron complex outermembrane recepter protein OS=Parasphingorhabdus marina DSM 22363 OX=1123272 GN=SAMN02745824_0327 PE=3 SV=1
MM1 pKa = 7.3SFSLSAMLLLAAPMAAEE18 pKa = 3.83PLVPEE23 pKa = 4.52EE24 pKa = 4.45GARR27 pKa = 11.84SEE29 pKa = 4.97GIFVPPATWRR39 pKa = 11.84WAQVRR44 pKa = 11.84FRR46 pKa = 11.84KK47 pKa = 9.51RR48 pKa = 11.84VIIRR52 pKa = 11.84VPRR55 pKa = 11.84QRR57 pKa = 11.84PTRR60 pKa = 11.84PSTRR64 pKa = 11.84ASEE67 pKa = 4.42SRR69 pKa = 11.84TSAPLTYY76 pKa = 9.49RR77 pKa = 11.84EE78 pKa = 3.96KK79 pKa = 11.15RR80 pKa = 11.84MGKK83 pKa = 9.41CMPMNNILGVQMFGGRR99 pKa = 11.84NLDD102 pKa = 4.13LITRR106 pKa = 11.84DD107 pKa = 3.31RR108 pKa = 11.84KK109 pKa = 9.64RR110 pKa = 11.84VRR112 pKa = 11.84ARR114 pKa = 11.84LEE116 pKa = 4.1KK117 pKa = 10.25KK118 pKa = 9.16CQARR122 pKa = 11.84SFYY125 pKa = 10.97SGFYY129 pKa = 9.05MEE131 pKa = 5.09KK132 pKa = 10.3SEE134 pKa = 5.29DD135 pKa = 3.53GKK137 pKa = 10.78ICADD141 pKa = 3.75RR142 pKa = 11.84DD143 pKa = 3.64ILHH146 pKa = 6.64SRR148 pKa = 11.84TGSKK152 pKa = 10.8CEE154 pKa = 3.46IDD156 pKa = 3.36RR157 pKa = 11.84FRR159 pKa = 11.84EE160 pKa = 4.1LVPEE164 pKa = 4.27
MM1 pKa = 7.3SFSLSAMLLLAAPMAAEE18 pKa = 3.83PLVPEE23 pKa = 4.52EE24 pKa = 4.45GARR27 pKa = 11.84SEE29 pKa = 4.97GIFVPPATWRR39 pKa = 11.84WAQVRR44 pKa = 11.84FRR46 pKa = 11.84KK47 pKa = 9.51RR48 pKa = 11.84VIIRR52 pKa = 11.84VPRR55 pKa = 11.84QRR57 pKa = 11.84PTRR60 pKa = 11.84PSTRR64 pKa = 11.84ASEE67 pKa = 4.42SRR69 pKa = 11.84TSAPLTYY76 pKa = 9.49RR77 pKa = 11.84EE78 pKa = 3.96KK79 pKa = 11.15RR80 pKa = 11.84MGKK83 pKa = 9.41CMPMNNILGVQMFGGRR99 pKa = 11.84NLDD102 pKa = 4.13LITRR106 pKa = 11.84DD107 pKa = 3.31RR108 pKa = 11.84KK109 pKa = 9.64RR110 pKa = 11.84VRR112 pKa = 11.84ARR114 pKa = 11.84LEE116 pKa = 4.1KK117 pKa = 10.25KK118 pKa = 9.16CQARR122 pKa = 11.84SFYY125 pKa = 10.97SGFYY129 pKa = 9.05MEE131 pKa = 5.09KK132 pKa = 10.3SEE134 pKa = 5.29DD135 pKa = 3.53GKK137 pKa = 10.78ICADD141 pKa = 3.75RR142 pKa = 11.84DD143 pKa = 3.64ILHH146 pKa = 6.64SRR148 pKa = 11.84TGSKK152 pKa = 10.8CEE154 pKa = 3.46IDD156 pKa = 3.36RR157 pKa = 11.84FRR159 pKa = 11.84EE160 pKa = 4.1LVPEE164 pKa = 4.27
Molecular weight: 18.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1084313 |
39 |
5177 |
319.1 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.133 ± 0.058 | 0.821 ± 0.014 |
6.362 ± 0.03 | 6.17 ± 0.048 |
3.903 ± 0.033 | 8.557 ± 0.068 |
1.932 ± 0.024 | 5.736 ± 0.038 |
3.798 ± 0.041 | 9.499 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.636 ± 0.024 | 3.204 ± 0.032 |
4.922 ± 0.041 | 3.421 ± 0.021 |
6.225 ± 0.044 | 5.984 ± 0.039 |
5.254 ± 0.052 | 6.724 ± 0.037 |
1.384 ± 0.019 | 2.336 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
