
Hubei tombus-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.82
Get precalculated fractions of proteins
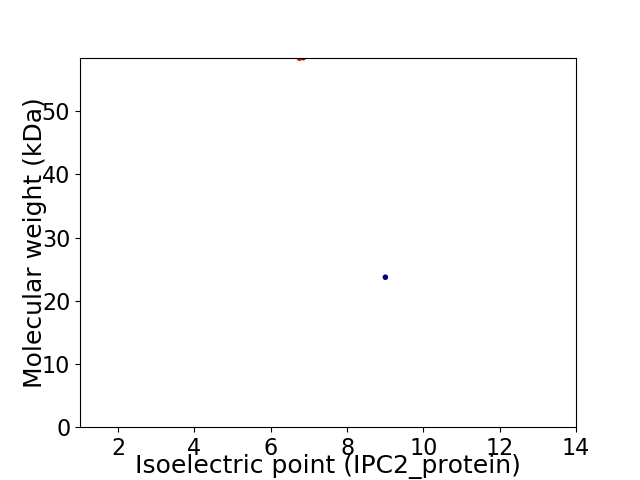
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
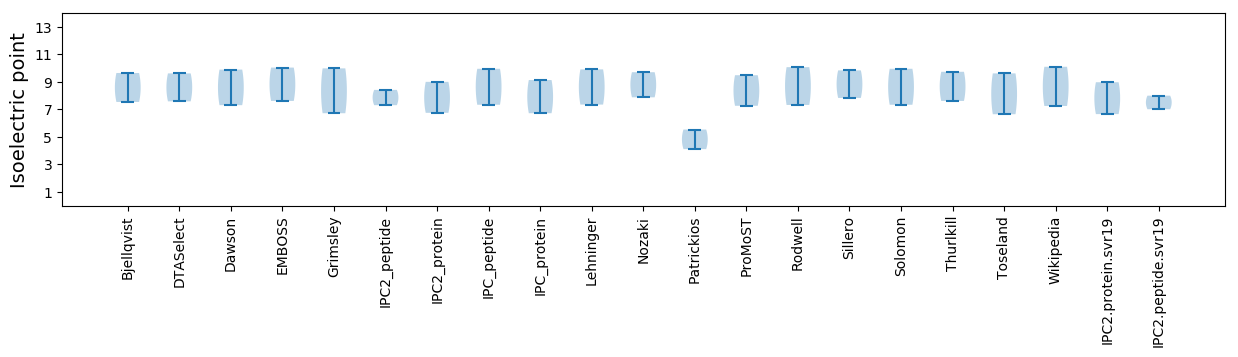
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGX1|A0A1L3KGX1_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 12 OX=1923258 PE=4 SV=1
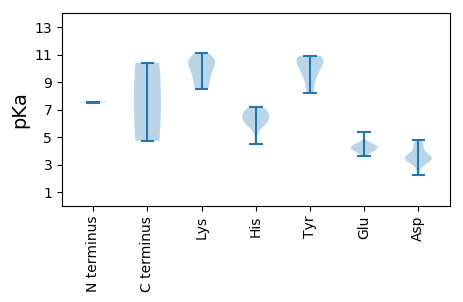
MM1 pKa = 7.51EE2 pKa = 5.38KK3 pKa = 10.54ADD5 pKa = 4.6DD6 pKa = 3.56RR7 pKa = 11.84HH8 pKa = 5.38GTPRR12 pKa = 11.84PPASKK17 pKa = 9.17GCEE20 pKa = 3.61SHH22 pKa = 6.69RR23 pKa = 11.84RR24 pKa = 11.84LIRR27 pKa = 11.84IVPPIYY33 pKa = 10.57GLWHH37 pKa = 6.97CFTHH41 pKa = 6.79SNCVCNDD48 pKa = 4.52LIACSNRR55 pKa = 11.84VIGVVPAPTDD65 pKa = 3.22TGIKK69 pKa = 9.86RR70 pKa = 11.84LRR72 pKa = 11.84EE73 pKa = 4.01AMNQLWPYY81 pKa = 10.86RR82 pKa = 11.84AMTPLSLEE90 pKa = 3.71QSLASFKK97 pKa = 10.79GSKK100 pKa = 9.94LKK102 pKa = 10.66LYY104 pKa = 10.0QRR106 pKa = 11.84AYY108 pKa = 10.76DD109 pKa = 3.79SLLITPLNQRR119 pKa = 11.84DD120 pKa = 3.7GRR122 pKa = 11.84VKK124 pKa = 10.88AFVKK128 pKa = 10.59AEE130 pKa = 4.08KK131 pKa = 10.05FDD133 pKa = 3.63PTEE136 pKa = 4.19KK137 pKa = 10.63VNPDD141 pKa = 2.77PRR143 pKa = 11.84MIQARR148 pKa = 11.84DD149 pKa = 3.27PRR151 pKa = 11.84YY152 pKa = 9.76NLHH155 pKa = 6.52LAQYY159 pKa = 9.39LRR161 pKa = 11.84PLEE164 pKa = 4.28HH165 pKa = 6.59EE166 pKa = 4.63CYY168 pKa = 10.25SLRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 6.08GAPAIAKK180 pKa = 8.56CFNPVQRR187 pKa = 11.84ANAIMQKK194 pKa = 9.83WNMFEE199 pKa = 4.5NPVCFSLDD207 pKa = 3.17CSRR210 pKa = 11.84WDD212 pKa = 3.23KK213 pKa = 10.86HH214 pKa = 4.52VHH216 pKa = 5.8RR217 pKa = 11.84KK218 pKa = 9.14VLDD221 pKa = 3.72VEE223 pKa = 4.51HH224 pKa = 6.98EE225 pKa = 4.98FYY227 pKa = 10.85QRR229 pKa = 11.84WYY231 pKa = 8.43PGEE234 pKa = 4.17VQLEE238 pKa = 4.42TLLGMQKK245 pKa = 10.49VNICTTSNGVKK256 pKa = 9.07YY257 pKa = 8.65TVDD260 pKa = 3.54GGRR263 pKa = 11.84MSGDD267 pKa = 3.19MNTALGNVTLMCGMIYY283 pKa = 10.67GAMKK287 pKa = 10.31TFDD290 pKa = 3.42NCHH293 pKa = 6.28FEE295 pKa = 4.01WLDD298 pKa = 4.4DD299 pKa = 4.47GDD301 pKa = 4.18DD302 pKa = 3.63CLVFVEE308 pKa = 4.14QHH310 pKa = 6.04DD311 pKa = 4.3LARR314 pKa = 11.84VAADD318 pKa = 3.79LPRR321 pKa = 11.84LFLEE325 pKa = 4.52YY326 pKa = 10.52GQEE329 pKa = 3.91LKK331 pKa = 10.56IEE333 pKa = 4.53NIAMNPQDD341 pKa = 4.76IVFCQSKK348 pKa = 8.47LTYY351 pKa = 10.3NGQFWTMARR360 pKa = 11.84NWRR363 pKa = 11.84KK364 pKa = 9.47VLSQSCCGTKK374 pKa = 9.43HH375 pKa = 5.95WNDD378 pKa = 3.49PNMVPGMFGLIGDD391 pKa = 4.78CEE393 pKa = 4.3MALHH397 pKa = 6.7RR398 pKa = 11.84GIPILQAFAQRR409 pKa = 11.84LRR411 pKa = 11.84DD412 pKa = 3.49LSGGRR417 pKa = 11.84RR418 pKa = 11.84ARR420 pKa = 11.84MEE422 pKa = 4.19HH423 pKa = 6.41MDD425 pKa = 3.24SSFQYY430 pKa = 10.4RR431 pKa = 11.84VGSYY435 pKa = 9.38QLGDD439 pKa = 3.4IQSITPAEE447 pKa = 4.23VTEE450 pKa = 4.01EE451 pKa = 3.82ARR453 pKa = 11.84FEE455 pKa = 4.14FQRR458 pKa = 11.84TWGVDD463 pKa = 2.64IQTQIGIEE471 pKa = 3.71WHH473 pKa = 5.99LAQWTPGIVHH483 pKa = 7.1RR484 pKa = 11.84DD485 pKa = 3.33VGPEE489 pKa = 3.87LFSTDD494 pKa = 2.23WCQQLEE500 pKa = 4.27PGIPNPTVLL509 pKa = 4.72
MM1 pKa = 7.51EE2 pKa = 5.38KK3 pKa = 10.54ADD5 pKa = 4.6DD6 pKa = 3.56RR7 pKa = 11.84HH8 pKa = 5.38GTPRR12 pKa = 11.84PPASKK17 pKa = 9.17GCEE20 pKa = 3.61SHH22 pKa = 6.69RR23 pKa = 11.84RR24 pKa = 11.84LIRR27 pKa = 11.84IVPPIYY33 pKa = 10.57GLWHH37 pKa = 6.97CFTHH41 pKa = 6.79SNCVCNDD48 pKa = 4.52LIACSNRR55 pKa = 11.84VIGVVPAPTDD65 pKa = 3.22TGIKK69 pKa = 9.86RR70 pKa = 11.84LRR72 pKa = 11.84EE73 pKa = 4.01AMNQLWPYY81 pKa = 10.86RR82 pKa = 11.84AMTPLSLEE90 pKa = 3.71QSLASFKK97 pKa = 10.79GSKK100 pKa = 9.94LKK102 pKa = 10.66LYY104 pKa = 10.0QRR106 pKa = 11.84AYY108 pKa = 10.76DD109 pKa = 3.79SLLITPLNQRR119 pKa = 11.84DD120 pKa = 3.7GRR122 pKa = 11.84VKK124 pKa = 10.88AFVKK128 pKa = 10.59AEE130 pKa = 4.08KK131 pKa = 10.05FDD133 pKa = 3.63PTEE136 pKa = 4.19KK137 pKa = 10.63VNPDD141 pKa = 2.77PRR143 pKa = 11.84MIQARR148 pKa = 11.84DD149 pKa = 3.27PRR151 pKa = 11.84YY152 pKa = 9.76NLHH155 pKa = 6.52LAQYY159 pKa = 9.39LRR161 pKa = 11.84PLEE164 pKa = 4.28HH165 pKa = 6.59EE166 pKa = 4.63CYY168 pKa = 10.25SLRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 6.08GAPAIAKK180 pKa = 8.56CFNPVQRR187 pKa = 11.84ANAIMQKK194 pKa = 9.83WNMFEE199 pKa = 4.5NPVCFSLDD207 pKa = 3.17CSRR210 pKa = 11.84WDD212 pKa = 3.23KK213 pKa = 10.86HH214 pKa = 4.52VHH216 pKa = 5.8RR217 pKa = 11.84KK218 pKa = 9.14VLDD221 pKa = 3.72VEE223 pKa = 4.51HH224 pKa = 6.98EE225 pKa = 4.98FYY227 pKa = 10.85QRR229 pKa = 11.84WYY231 pKa = 8.43PGEE234 pKa = 4.17VQLEE238 pKa = 4.42TLLGMQKK245 pKa = 10.49VNICTTSNGVKK256 pKa = 9.07YY257 pKa = 8.65TVDD260 pKa = 3.54GGRR263 pKa = 11.84MSGDD267 pKa = 3.19MNTALGNVTLMCGMIYY283 pKa = 10.67GAMKK287 pKa = 10.31TFDD290 pKa = 3.42NCHH293 pKa = 6.28FEE295 pKa = 4.01WLDD298 pKa = 4.4DD299 pKa = 4.47GDD301 pKa = 4.18DD302 pKa = 3.63CLVFVEE308 pKa = 4.14QHH310 pKa = 6.04DD311 pKa = 4.3LARR314 pKa = 11.84VAADD318 pKa = 3.79LPRR321 pKa = 11.84LFLEE325 pKa = 4.52YY326 pKa = 10.52GQEE329 pKa = 3.91LKK331 pKa = 10.56IEE333 pKa = 4.53NIAMNPQDD341 pKa = 4.76IVFCQSKK348 pKa = 8.47LTYY351 pKa = 10.3NGQFWTMARR360 pKa = 11.84NWRR363 pKa = 11.84KK364 pKa = 9.47VLSQSCCGTKK374 pKa = 9.43HH375 pKa = 5.95WNDD378 pKa = 3.49PNMVPGMFGLIGDD391 pKa = 4.78CEE393 pKa = 4.3MALHH397 pKa = 6.7RR398 pKa = 11.84GIPILQAFAQRR409 pKa = 11.84LRR411 pKa = 11.84DD412 pKa = 3.49LSGGRR417 pKa = 11.84RR418 pKa = 11.84ARR420 pKa = 11.84MEE422 pKa = 4.19HH423 pKa = 6.41MDD425 pKa = 3.24SSFQYY430 pKa = 10.4RR431 pKa = 11.84VGSYY435 pKa = 9.38QLGDD439 pKa = 3.4IQSITPAEE447 pKa = 4.23VTEE450 pKa = 4.01EE451 pKa = 3.82ARR453 pKa = 11.84FEE455 pKa = 4.14FQRR458 pKa = 11.84TWGVDD463 pKa = 2.64IQTQIGIEE471 pKa = 3.71WHH473 pKa = 5.99LAQWTPGIVHH483 pKa = 7.1RR484 pKa = 11.84DD485 pKa = 3.33VGPEE489 pKa = 3.87LFSTDD494 pKa = 2.23WCQQLEE500 pKa = 4.27PGIPNPTVLL509 pKa = 4.72
Molecular weight: 58.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGX1|A0A1L3KGX1_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 12 OX=1923258 PE=4 SV=1
MM1 pKa = 7.57SKK3 pKa = 8.49PTRR6 pKa = 11.84RR7 pKa = 11.84THH9 pKa = 6.3WLPTATYY16 pKa = 9.42RR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84GSKK23 pKa = 8.48TPSGPSSVASDD34 pKa = 4.08LPSTDD39 pKa = 3.16EE40 pKa = 4.34LRR42 pKa = 11.84SEE44 pKa = 4.51LSRR47 pKa = 11.84LEE49 pKa = 4.28EE50 pKa = 4.13EE51 pKa = 4.35AQKK54 pKa = 9.85VQPWTDD60 pKa = 3.15VEE62 pKa = 4.25VRR64 pKa = 11.84LLGFLRR70 pKa = 11.84GRR72 pKa = 11.84LYY74 pKa = 10.64GAKK77 pKa = 8.67RR78 pKa = 11.84TPATPQSLNLQGLNWCVKK96 pKa = 9.46NHH98 pKa = 6.1DD99 pKa = 4.14HH100 pKa = 6.73LGEE103 pKa = 4.19YY104 pKa = 8.2QHH106 pKa = 7.15PAAQAALVARR116 pKa = 11.84CVNVVLPPSLEE127 pKa = 3.93EE128 pKa = 4.33AGLIDD133 pKa = 3.74VYY135 pKa = 10.9QRR137 pKa = 11.84EE138 pKa = 4.49GEE140 pKa = 4.19KK141 pKa = 10.29IKK143 pKa = 11.1AFNNGLDD150 pKa = 3.48EE151 pKa = 4.46MGDD154 pKa = 3.37RR155 pKa = 11.84RR156 pKa = 11.84EE157 pKa = 3.97DD158 pKa = 3.66LRR160 pKa = 11.84WLVGLGGVTAAVAGTVAGLTGSRR183 pKa = 11.84VIGGGLLAAGIAGVGIYY200 pKa = 9.79GWSKK204 pKa = 10.26VRR206 pKa = 11.84DD207 pKa = 3.42ITRR210 pKa = 11.84RR211 pKa = 11.84AQAARR216 pKa = 11.84WYY218 pKa = 10.41
MM1 pKa = 7.57SKK3 pKa = 8.49PTRR6 pKa = 11.84RR7 pKa = 11.84THH9 pKa = 6.3WLPTATYY16 pKa = 9.42RR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84GSKK23 pKa = 8.48TPSGPSSVASDD34 pKa = 4.08LPSTDD39 pKa = 3.16EE40 pKa = 4.34LRR42 pKa = 11.84SEE44 pKa = 4.51LSRR47 pKa = 11.84LEE49 pKa = 4.28EE50 pKa = 4.13EE51 pKa = 4.35AQKK54 pKa = 9.85VQPWTDD60 pKa = 3.15VEE62 pKa = 4.25VRR64 pKa = 11.84LLGFLRR70 pKa = 11.84GRR72 pKa = 11.84LYY74 pKa = 10.64GAKK77 pKa = 8.67RR78 pKa = 11.84TPATPQSLNLQGLNWCVKK96 pKa = 9.46NHH98 pKa = 6.1DD99 pKa = 4.14HH100 pKa = 6.73LGEE103 pKa = 4.19YY104 pKa = 8.2QHH106 pKa = 7.15PAAQAALVARR116 pKa = 11.84CVNVVLPPSLEE127 pKa = 3.93EE128 pKa = 4.33AGLIDD133 pKa = 3.74VYY135 pKa = 10.9QRR137 pKa = 11.84EE138 pKa = 4.49GEE140 pKa = 4.19KK141 pKa = 10.29IKK143 pKa = 11.1AFNNGLDD150 pKa = 3.48EE151 pKa = 4.46MGDD154 pKa = 3.37RR155 pKa = 11.84RR156 pKa = 11.84EE157 pKa = 3.97DD158 pKa = 3.66LRR160 pKa = 11.84WLVGLGGVTAAVAGTVAGLTGSRR183 pKa = 11.84VIGGGLLAAGIAGVGIYY200 pKa = 9.79GWSKK204 pKa = 10.26VRR206 pKa = 11.84DD207 pKa = 3.42ITRR210 pKa = 11.84RR211 pKa = 11.84AQAARR216 pKa = 11.84WYY218 pKa = 10.41
Molecular weight: 23.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
727 |
218 |
509 |
363.5 |
41.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.29 ± 1.398 | 2.751 ± 0.915 |
5.365 ± 0.617 | 5.502 ± 0.23 |
3.026 ± 1.052 | 8.116 ± 1.672 |
2.889 ± 0.526 | 4.264 ± 0.754 |
3.989 ± 0.159 | 9.354 ± 0.826 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.889 ± 0.983 | 3.989 ± 0.617 |
5.64 ± 0.296 | 4.952 ± 0.64 |
7.978 ± 0.826 | 4.952 ± 0.505 |
5.089 ± 0.436 | 6.465 ± 0.665 |
2.613 ± 0.069 | 2.889 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
