
Phytophthora infestans RNA virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
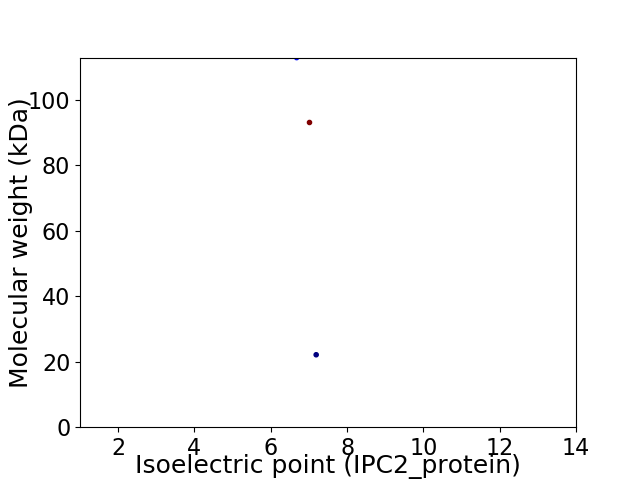
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
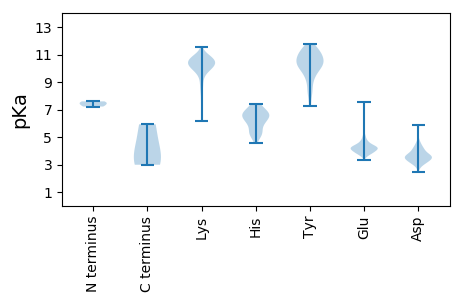
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7SHT6|C7SHT6_9VIRU Uncharacterized protein ORF2A OS=Phytophthora infestans RNA virus 1 OX=640897 GN=ORF2A PE=4 SV=1
MM1 pKa = 7.22QINVGGRR8 pKa = 11.84EE9 pKa = 3.7LCAIFVGLICWMFICEE25 pKa = 4.76FIVNEE30 pKa = 3.97YY31 pKa = 9.04EE32 pKa = 3.89QFYY35 pKa = 8.08GTEE38 pKa = 4.32YY39 pKa = 10.35TVEE42 pKa = 4.12TSPMCALFEE51 pKa = 4.47GCSLNVEE58 pKa = 4.26SFLKK62 pKa = 11.06NEE64 pKa = 4.26DD65 pKa = 3.15VRR67 pKa = 11.84SIMITALFVGKK78 pKa = 8.9MLEE81 pKa = 4.1MLDD84 pKa = 3.21ITVKK88 pKa = 10.86FEE90 pKa = 3.84FSSAEE95 pKa = 3.76EE96 pKa = 4.54GEE98 pKa = 4.87SVFADD103 pKa = 4.54FNRR106 pKa = 11.84DD107 pKa = 3.05LRR109 pKa = 11.84DD110 pKa = 3.17VRR112 pKa = 11.84RR113 pKa = 11.84SVEE116 pKa = 4.06LKK118 pKa = 9.83ASSMKK123 pKa = 10.49KK124 pKa = 9.79IIADD128 pKa = 3.66FRR130 pKa = 11.84TKK132 pKa = 10.01IRR134 pKa = 11.84SIVYY138 pKa = 10.01LGVSNIDD145 pKa = 3.31IPNVHH150 pKa = 6.84AMRR153 pKa = 11.84YY154 pKa = 9.06PIDD157 pKa = 3.63FFMSRR162 pKa = 11.84LYY164 pKa = 10.87RR165 pKa = 11.84MKK167 pKa = 10.19FLKK170 pKa = 10.26EE171 pKa = 3.92YY172 pKa = 10.24EE173 pKa = 4.16EE174 pKa = 4.12SRR176 pKa = 11.84EE177 pKa = 3.79KK178 pKa = 10.56YY179 pKa = 9.96RR180 pKa = 11.84RR181 pKa = 11.84VNPTLAMKK189 pKa = 10.21LAQLLRR195 pKa = 11.84YY196 pKa = 10.05DD197 pKa = 4.02NEE199 pKa = 4.29FLLEE203 pKa = 4.28SNVAEE208 pKa = 4.05ALKK211 pKa = 10.71EE212 pKa = 4.16SVEE215 pKa = 4.22EE216 pKa = 3.87VLEE219 pKa = 4.3EE220 pKa = 4.3VFSNPSFEE228 pKa = 4.16EE229 pKa = 3.86VKK231 pKa = 10.48IEE233 pKa = 4.38LNGRR237 pKa = 11.84LNSGMAMFDD246 pKa = 4.23DD247 pKa = 4.21INVNGSSAAGYY258 pKa = 7.42PYY260 pKa = 9.8PAGVKK265 pKa = 9.99RR266 pKa = 11.84RR267 pKa = 11.84DD268 pKa = 3.48VLEE271 pKa = 3.99EE272 pKa = 4.1AKK274 pKa = 10.8ASAYY278 pKa = 10.4DD279 pKa = 4.13LYY281 pKa = 11.42NDD283 pKa = 3.39SDD285 pKa = 4.02LFEE288 pKa = 7.53GYY290 pKa = 9.63MSDD293 pKa = 3.48HH294 pKa = 6.57RR295 pKa = 11.84WYY297 pKa = 7.24TTGRR301 pKa = 11.84AKK303 pKa = 10.13LVKK306 pKa = 9.42MGKK309 pKa = 8.53PDD311 pKa = 3.46KK312 pKa = 10.87ARR314 pKa = 11.84LVLYY318 pKa = 10.82SGFSYY323 pKa = 11.15SLLGFVYY330 pKa = 10.05SQVWTGFMNRR340 pKa = 11.84QCRR343 pKa = 11.84GWSAVGMSWMNGGAAKK359 pKa = 10.17VASFFEE365 pKa = 4.65DD366 pKa = 3.62CFGIAVSGFEE376 pKa = 4.46YY377 pKa = 8.23MTLDD381 pKa = 3.29VAEE384 pKa = 4.58WDD386 pKa = 3.61SSVCRR391 pKa = 11.84EE392 pKa = 4.46LLHH395 pKa = 5.85ACKK398 pKa = 9.83RR399 pKa = 11.84FHH401 pKa = 6.16MRR403 pKa = 11.84VLEE406 pKa = 4.14RR407 pKa = 11.84TLSPEE412 pKa = 3.36NARR415 pKa = 11.84YY416 pKa = 9.53KK417 pKa = 10.56EE418 pKa = 4.14YY419 pKa = 10.52FGRR422 pKa = 11.84IYY424 pKa = 11.05DD425 pKa = 3.74EE426 pKa = 4.38MIEE429 pKa = 4.25AKK431 pKa = 10.25VVLPGGHH438 pKa = 5.95SFRR441 pKa = 11.84LHH443 pKa = 7.09HH444 pKa = 6.12GMKK447 pKa = 10.21SGWIMTANDD456 pKa = 3.55NTLMHH461 pKa = 6.56EE462 pKa = 4.36FVVRR466 pKa = 11.84TLQKK470 pKa = 10.04IGQIPDD476 pKa = 3.52MKK478 pKa = 10.2RR479 pKa = 11.84QLYY482 pKa = 10.78GDD484 pKa = 4.78DD485 pKa = 3.51NLSRR489 pKa = 11.84KK490 pKa = 8.97PIGMSKK496 pKa = 9.94QLLVDD501 pKa = 4.25GYY503 pKa = 11.63GMFGFRR509 pKa = 11.84LSHH512 pKa = 5.33IHH514 pKa = 4.54VSRR517 pKa = 11.84RR518 pKa = 11.84LSEE521 pKa = 3.97VDD523 pKa = 4.03FLSKK527 pKa = 10.31FIIFKK532 pKa = 10.69DD533 pKa = 3.4GFYY536 pKa = 10.64FPWRR540 pKa = 11.84EE541 pKa = 3.7QTEE544 pKa = 4.17THH546 pKa = 6.22ARR548 pKa = 11.84LLMPEE553 pKa = 4.28EE554 pKa = 4.16FDD556 pKa = 3.47PNYY559 pKa = 10.54RR560 pKa = 11.84IVPDD564 pKa = 3.39SRR566 pKa = 11.84VAAEE570 pKa = 4.32HH571 pKa = 6.64LLGHH575 pKa = 7.32LLDD578 pKa = 4.58NPFNANVRR586 pKa = 11.84HH587 pKa = 5.16VCFTLLEE594 pKa = 4.43YY595 pKa = 10.39IKK597 pKa = 10.82SNYY600 pKa = 8.21GVRR603 pKa = 11.84TINVEE608 pKa = 3.84EE609 pKa = 4.71FINTRR614 pKa = 11.84WSFMLRR620 pKa = 11.84DD621 pKa = 3.82IKK623 pKa = 10.62RR624 pKa = 11.84LCGEE628 pKa = 4.02IPTVPSVEE636 pKa = 5.06FIQEE640 pKa = 4.14LYY642 pKa = 11.08GVFSEE647 pKa = 4.28PVSFNWAGVGEE658 pKa = 4.44YY659 pKa = 10.32ARR661 pKa = 11.84KK662 pKa = 9.58LPKK665 pKa = 10.15FSFISKK671 pKa = 10.12APSAEE676 pKa = 3.85PFFRR680 pKa = 11.84SQKK683 pKa = 10.52AAFEE687 pKa = 4.43LSVSEE692 pKa = 4.3KK693 pKa = 10.09FGHH696 pKa = 6.58RR697 pKa = 11.84KK698 pKa = 7.53TSALNRR704 pKa = 11.84ACGYY708 pKa = 10.91KK709 pKa = 10.31KK710 pKa = 9.95MPLHH714 pKa = 6.87IFGQAGGKK722 pKa = 9.62LIEE725 pKa = 5.17IINTCGLKK733 pKa = 10.53CEE735 pKa = 4.33SVLDD739 pKa = 4.51LGTHH743 pKa = 7.07PGAAMATLQSMHH755 pKa = 6.29KK756 pKa = 9.55VKK758 pKa = 10.77SATCVSLFPEE768 pKa = 4.06IDD770 pKa = 3.31KK771 pKa = 11.52SKK773 pKa = 10.43GFCPRR778 pKa = 11.84VLRR781 pKa = 11.84GKK783 pKa = 10.12DD784 pKa = 3.37FEE786 pKa = 4.22FFEE789 pKa = 4.44MSAEE793 pKa = 3.97KK794 pKa = 10.33FKK796 pKa = 9.59PTKK799 pKa = 10.34HH800 pKa = 6.68YY801 pKa = 11.32DD802 pKa = 3.0MCFDD806 pKa = 5.88DD807 pKa = 5.78IYY809 pKa = 11.49DD810 pKa = 3.62FEE812 pKa = 4.24EE813 pKa = 4.41ADD815 pKa = 3.42RR816 pKa = 11.84FKK818 pKa = 10.8RR819 pKa = 11.84AEE821 pKa = 3.77QMFGHH826 pKa = 6.62VDD828 pKa = 2.98RR829 pKa = 11.84HH830 pKa = 5.06FAGALTRR837 pKa = 11.84AEE839 pKa = 4.23KK840 pKa = 10.41FKK842 pKa = 10.2PFCGSYY848 pKa = 9.33VMKK851 pKa = 10.77VRR853 pKa = 11.84GFTPLVLKK861 pKa = 10.54GLYY864 pKa = 9.79DD865 pKa = 4.3LYY867 pKa = 11.55RR868 pKa = 11.84MWGYY872 pKa = 11.38LDD874 pKa = 3.16IRR876 pKa = 11.84KK877 pKa = 7.53TFYY880 pKa = 11.02SNPWNPEE887 pKa = 3.7FYY889 pKa = 9.96VVLVKK894 pKa = 10.73RR895 pKa = 11.84HH896 pKa = 5.14DD897 pKa = 3.48QYY899 pKa = 11.8VRR901 pKa = 11.84RR902 pKa = 11.84STFNGAVNAYY912 pKa = 10.1LNGIAPRR919 pKa = 11.84LSFFVNARR927 pKa = 11.84AFNYY931 pKa = 10.54DD932 pKa = 3.04RR933 pKa = 11.84MSLGRR938 pKa = 11.84EE939 pKa = 3.91TLNNPLRR946 pKa = 11.84DD947 pKa = 3.84DD948 pKa = 3.75EE949 pKa = 4.64TVQRR953 pKa = 11.84AFDD956 pKa = 3.97AYY958 pKa = 10.82AEE960 pKa = 4.23TLRR963 pKa = 11.84AQSFNDD969 pKa = 3.85DD970 pKa = 3.8YY971 pKa = 10.29PTEE974 pKa = 4.14VMFF977 pKa = 5.93
MM1 pKa = 7.22QINVGGRR8 pKa = 11.84EE9 pKa = 3.7LCAIFVGLICWMFICEE25 pKa = 4.76FIVNEE30 pKa = 3.97YY31 pKa = 9.04EE32 pKa = 3.89QFYY35 pKa = 8.08GTEE38 pKa = 4.32YY39 pKa = 10.35TVEE42 pKa = 4.12TSPMCALFEE51 pKa = 4.47GCSLNVEE58 pKa = 4.26SFLKK62 pKa = 11.06NEE64 pKa = 4.26DD65 pKa = 3.15VRR67 pKa = 11.84SIMITALFVGKK78 pKa = 8.9MLEE81 pKa = 4.1MLDD84 pKa = 3.21ITVKK88 pKa = 10.86FEE90 pKa = 3.84FSSAEE95 pKa = 3.76EE96 pKa = 4.54GEE98 pKa = 4.87SVFADD103 pKa = 4.54FNRR106 pKa = 11.84DD107 pKa = 3.05LRR109 pKa = 11.84DD110 pKa = 3.17VRR112 pKa = 11.84RR113 pKa = 11.84SVEE116 pKa = 4.06LKK118 pKa = 9.83ASSMKK123 pKa = 10.49KK124 pKa = 9.79IIADD128 pKa = 3.66FRR130 pKa = 11.84TKK132 pKa = 10.01IRR134 pKa = 11.84SIVYY138 pKa = 10.01LGVSNIDD145 pKa = 3.31IPNVHH150 pKa = 6.84AMRR153 pKa = 11.84YY154 pKa = 9.06PIDD157 pKa = 3.63FFMSRR162 pKa = 11.84LYY164 pKa = 10.87RR165 pKa = 11.84MKK167 pKa = 10.19FLKK170 pKa = 10.26EE171 pKa = 3.92YY172 pKa = 10.24EE173 pKa = 4.16EE174 pKa = 4.12SRR176 pKa = 11.84EE177 pKa = 3.79KK178 pKa = 10.56YY179 pKa = 9.96RR180 pKa = 11.84RR181 pKa = 11.84VNPTLAMKK189 pKa = 10.21LAQLLRR195 pKa = 11.84YY196 pKa = 10.05DD197 pKa = 4.02NEE199 pKa = 4.29FLLEE203 pKa = 4.28SNVAEE208 pKa = 4.05ALKK211 pKa = 10.71EE212 pKa = 4.16SVEE215 pKa = 4.22EE216 pKa = 3.87VLEE219 pKa = 4.3EE220 pKa = 4.3VFSNPSFEE228 pKa = 4.16EE229 pKa = 3.86VKK231 pKa = 10.48IEE233 pKa = 4.38LNGRR237 pKa = 11.84LNSGMAMFDD246 pKa = 4.23DD247 pKa = 4.21INVNGSSAAGYY258 pKa = 7.42PYY260 pKa = 9.8PAGVKK265 pKa = 9.99RR266 pKa = 11.84RR267 pKa = 11.84DD268 pKa = 3.48VLEE271 pKa = 3.99EE272 pKa = 4.1AKK274 pKa = 10.8ASAYY278 pKa = 10.4DD279 pKa = 4.13LYY281 pKa = 11.42NDD283 pKa = 3.39SDD285 pKa = 4.02LFEE288 pKa = 7.53GYY290 pKa = 9.63MSDD293 pKa = 3.48HH294 pKa = 6.57RR295 pKa = 11.84WYY297 pKa = 7.24TTGRR301 pKa = 11.84AKK303 pKa = 10.13LVKK306 pKa = 9.42MGKK309 pKa = 8.53PDD311 pKa = 3.46KK312 pKa = 10.87ARR314 pKa = 11.84LVLYY318 pKa = 10.82SGFSYY323 pKa = 11.15SLLGFVYY330 pKa = 10.05SQVWTGFMNRR340 pKa = 11.84QCRR343 pKa = 11.84GWSAVGMSWMNGGAAKK359 pKa = 10.17VASFFEE365 pKa = 4.65DD366 pKa = 3.62CFGIAVSGFEE376 pKa = 4.46YY377 pKa = 8.23MTLDD381 pKa = 3.29VAEE384 pKa = 4.58WDD386 pKa = 3.61SSVCRR391 pKa = 11.84EE392 pKa = 4.46LLHH395 pKa = 5.85ACKK398 pKa = 9.83RR399 pKa = 11.84FHH401 pKa = 6.16MRR403 pKa = 11.84VLEE406 pKa = 4.14RR407 pKa = 11.84TLSPEE412 pKa = 3.36NARR415 pKa = 11.84YY416 pKa = 9.53KK417 pKa = 10.56EE418 pKa = 4.14YY419 pKa = 10.52FGRR422 pKa = 11.84IYY424 pKa = 11.05DD425 pKa = 3.74EE426 pKa = 4.38MIEE429 pKa = 4.25AKK431 pKa = 10.25VVLPGGHH438 pKa = 5.95SFRR441 pKa = 11.84LHH443 pKa = 7.09HH444 pKa = 6.12GMKK447 pKa = 10.21SGWIMTANDD456 pKa = 3.55NTLMHH461 pKa = 6.56EE462 pKa = 4.36FVVRR466 pKa = 11.84TLQKK470 pKa = 10.04IGQIPDD476 pKa = 3.52MKK478 pKa = 10.2RR479 pKa = 11.84QLYY482 pKa = 10.78GDD484 pKa = 4.78DD485 pKa = 3.51NLSRR489 pKa = 11.84KK490 pKa = 8.97PIGMSKK496 pKa = 9.94QLLVDD501 pKa = 4.25GYY503 pKa = 11.63GMFGFRR509 pKa = 11.84LSHH512 pKa = 5.33IHH514 pKa = 4.54VSRR517 pKa = 11.84RR518 pKa = 11.84LSEE521 pKa = 3.97VDD523 pKa = 4.03FLSKK527 pKa = 10.31FIIFKK532 pKa = 10.69DD533 pKa = 3.4GFYY536 pKa = 10.64FPWRR540 pKa = 11.84EE541 pKa = 3.7QTEE544 pKa = 4.17THH546 pKa = 6.22ARR548 pKa = 11.84LLMPEE553 pKa = 4.28EE554 pKa = 4.16FDD556 pKa = 3.47PNYY559 pKa = 10.54RR560 pKa = 11.84IVPDD564 pKa = 3.39SRR566 pKa = 11.84VAAEE570 pKa = 4.32HH571 pKa = 6.64LLGHH575 pKa = 7.32LLDD578 pKa = 4.58NPFNANVRR586 pKa = 11.84HH587 pKa = 5.16VCFTLLEE594 pKa = 4.43YY595 pKa = 10.39IKK597 pKa = 10.82SNYY600 pKa = 8.21GVRR603 pKa = 11.84TINVEE608 pKa = 3.84EE609 pKa = 4.71FINTRR614 pKa = 11.84WSFMLRR620 pKa = 11.84DD621 pKa = 3.82IKK623 pKa = 10.62RR624 pKa = 11.84LCGEE628 pKa = 4.02IPTVPSVEE636 pKa = 5.06FIQEE640 pKa = 4.14LYY642 pKa = 11.08GVFSEE647 pKa = 4.28PVSFNWAGVGEE658 pKa = 4.44YY659 pKa = 10.32ARR661 pKa = 11.84KK662 pKa = 9.58LPKK665 pKa = 10.15FSFISKK671 pKa = 10.12APSAEE676 pKa = 3.85PFFRR680 pKa = 11.84SQKK683 pKa = 10.52AAFEE687 pKa = 4.43LSVSEE692 pKa = 4.3KK693 pKa = 10.09FGHH696 pKa = 6.58RR697 pKa = 11.84KK698 pKa = 7.53TSALNRR704 pKa = 11.84ACGYY708 pKa = 10.91KK709 pKa = 10.31KK710 pKa = 9.95MPLHH714 pKa = 6.87IFGQAGGKK722 pKa = 9.62LIEE725 pKa = 5.17IINTCGLKK733 pKa = 10.53CEE735 pKa = 4.33SVLDD739 pKa = 4.51LGTHH743 pKa = 7.07PGAAMATLQSMHH755 pKa = 6.29KK756 pKa = 9.55VKK758 pKa = 10.77SATCVSLFPEE768 pKa = 4.06IDD770 pKa = 3.31KK771 pKa = 11.52SKK773 pKa = 10.43GFCPRR778 pKa = 11.84VLRR781 pKa = 11.84GKK783 pKa = 10.12DD784 pKa = 3.37FEE786 pKa = 4.22FFEE789 pKa = 4.44MSAEE793 pKa = 3.97KK794 pKa = 10.33FKK796 pKa = 9.59PTKK799 pKa = 10.34HH800 pKa = 6.68YY801 pKa = 11.32DD802 pKa = 3.0MCFDD806 pKa = 5.88DD807 pKa = 5.78IYY809 pKa = 11.49DD810 pKa = 3.62FEE812 pKa = 4.24EE813 pKa = 4.41ADD815 pKa = 3.42RR816 pKa = 11.84FKK818 pKa = 10.8RR819 pKa = 11.84AEE821 pKa = 3.77QMFGHH826 pKa = 6.62VDD828 pKa = 2.98RR829 pKa = 11.84HH830 pKa = 5.06FAGALTRR837 pKa = 11.84AEE839 pKa = 4.23KK840 pKa = 10.41FKK842 pKa = 10.2PFCGSYY848 pKa = 9.33VMKK851 pKa = 10.77VRR853 pKa = 11.84GFTPLVLKK861 pKa = 10.54GLYY864 pKa = 9.79DD865 pKa = 4.3LYY867 pKa = 11.55RR868 pKa = 11.84MWGYY872 pKa = 11.38LDD874 pKa = 3.16IRR876 pKa = 11.84KK877 pKa = 7.53TFYY880 pKa = 11.02SNPWNPEE887 pKa = 3.7FYY889 pKa = 9.96VVLVKK894 pKa = 10.73RR895 pKa = 11.84HH896 pKa = 5.14DD897 pKa = 3.48QYY899 pKa = 11.8VRR901 pKa = 11.84RR902 pKa = 11.84STFNGAVNAYY912 pKa = 10.1LNGIAPRR919 pKa = 11.84LSFFVNARR927 pKa = 11.84AFNYY931 pKa = 10.54DD932 pKa = 3.04RR933 pKa = 11.84MSLGRR938 pKa = 11.84EE939 pKa = 3.91TLNNPLRR946 pKa = 11.84DD947 pKa = 3.84DD948 pKa = 3.75EE949 pKa = 4.64TVQRR953 pKa = 11.84AFDD956 pKa = 3.97AYY958 pKa = 10.82AEE960 pKa = 4.23TLRR963 pKa = 11.84AQSFNDD969 pKa = 3.85DD970 pKa = 3.8YY971 pKa = 10.29PTEE974 pKa = 4.14VMFF977 pKa = 5.93
Molecular weight: 112.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7SHT7|C7SHT7_9VIRU Uncharacterized protein ORF3A OS=Phytophthora infestans RNA virus 1 OX=640897 GN=ORF3A PE=4 SV=1
MM1 pKa = 7.59VSEE4 pKa = 4.59VVLVDD9 pKa = 3.55YY10 pKa = 11.24SRR12 pKa = 11.84DD13 pKa = 3.16RR14 pKa = 11.84HH15 pKa = 5.28AVTRR19 pKa = 11.84AAQWVFSLPARR30 pKa = 11.84LDD32 pKa = 4.09GVCPWVRR39 pKa = 11.84EE40 pKa = 4.24EE41 pKa = 4.33EE42 pKa = 4.26CSLLCQVICFFRR54 pKa = 11.84WLAEE58 pKa = 3.7WKK60 pKa = 10.15RR61 pKa = 11.84WVACLLPSNEE71 pKa = 3.73VRR73 pKa = 11.84GMRR76 pKa = 11.84TEE78 pKa = 3.86IGVQEE83 pKa = 4.21AQLGVNSPINRR94 pKa = 11.84SWMEE98 pKa = 3.82LRR100 pKa = 11.84SLAVFVSVIVCGVILSVIGFLRR122 pKa = 11.84HH123 pKa = 5.31RR124 pKa = 11.84RR125 pKa = 11.84ALRR128 pKa = 11.84STRR131 pKa = 11.84LLTVMGLAQLLIVTIAGMTVTLTDD155 pKa = 3.93FLCVISLMEE164 pKa = 4.24LVTLEE169 pKa = 4.08MAVLLEE175 pKa = 4.1SFLEE179 pKa = 4.29FFEE182 pKa = 4.02MRR184 pKa = 11.84RR185 pKa = 11.84NRR187 pKa = 11.84LVPVSISS194 pKa = 2.98
MM1 pKa = 7.59VSEE4 pKa = 4.59VVLVDD9 pKa = 3.55YY10 pKa = 11.24SRR12 pKa = 11.84DD13 pKa = 3.16RR14 pKa = 11.84HH15 pKa = 5.28AVTRR19 pKa = 11.84AAQWVFSLPARR30 pKa = 11.84LDD32 pKa = 4.09GVCPWVRR39 pKa = 11.84EE40 pKa = 4.24EE41 pKa = 4.33EE42 pKa = 4.26CSLLCQVICFFRR54 pKa = 11.84WLAEE58 pKa = 3.7WKK60 pKa = 10.15RR61 pKa = 11.84WVACLLPSNEE71 pKa = 3.73VRR73 pKa = 11.84GMRR76 pKa = 11.84TEE78 pKa = 3.86IGVQEE83 pKa = 4.21AQLGVNSPINRR94 pKa = 11.84SWMEE98 pKa = 3.82LRR100 pKa = 11.84SLAVFVSVIVCGVILSVIGFLRR122 pKa = 11.84HH123 pKa = 5.31RR124 pKa = 11.84RR125 pKa = 11.84ALRR128 pKa = 11.84STRR131 pKa = 11.84LLTVMGLAQLLIVTIAGMTVTLTDD155 pKa = 3.93FLCVISLMEE164 pKa = 4.24LVTLEE169 pKa = 4.08MAVLLEE175 pKa = 4.1SFLEE179 pKa = 4.29FFEE182 pKa = 4.02MRR184 pKa = 11.84RR185 pKa = 11.84NRR187 pKa = 11.84LVPVSISS194 pKa = 2.98
Molecular weight: 22.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2018 |
194 |
977 |
672.7 |
75.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.938 ± 0.418 | 2.23 ± 0.316 |
4.559 ± 0.519 | 6.541 ± 0.856 |
7.433 ± 0.522 | 7.83 ± 1.459 |
1.685 ± 0.379 | 5.104 ± 0.494 |
4.609 ± 0.971 | 9.713 ± 1.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.072 ± 0.744 | 4.014 ± 0.37 |
2.924 ± 0.449 | 1.685 ± 0.236 |
6.739 ± 0.666 | 7.136 ± 0.14 |
3.865 ± 0.208 | 8.87 ± 1.391 |
1.437 ± 0.31 | 3.617 ± 0.717 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
