
Sanxia tombus-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.24
Get precalculated fractions of proteins
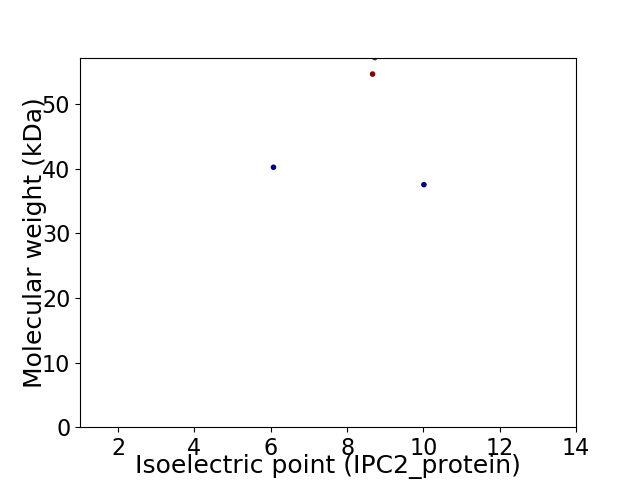
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGC2|A0A1L3KGC2_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 7 OX=1923391 PE=4 SV=1
MM1 pKa = 7.35TARR4 pKa = 11.84CVLAMPRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GTATNGAGYY22 pKa = 10.78APVSLGTGVRR32 pKa = 11.84ANRR35 pKa = 11.84RR36 pKa = 11.84TPPMQLEE43 pKa = 4.23GTDD46 pKa = 3.62RR47 pKa = 11.84IGVINWDD54 pKa = 3.47GALDD58 pKa = 3.78TGHH61 pKa = 6.92VLFDD65 pKa = 4.23LLVEE69 pKa = 4.2PGIFKK74 pKa = 10.6SLRR77 pKa = 11.84DD78 pKa = 3.52VAKK81 pKa = 10.41ARR83 pKa = 11.84QYY85 pKa = 9.87IQWHH89 pKa = 5.12ALEE92 pKa = 5.26FKK94 pKa = 10.25FSCGTNTAKK103 pKa = 10.76DD104 pKa = 3.84GSYY107 pKa = 10.38VAAFIADD114 pKa = 4.17PDD116 pKa = 4.58DD117 pKa = 5.0KK118 pKa = 11.76NPDD121 pKa = 3.76DD122 pKa = 3.69PTLAVEE128 pKa = 4.81EE129 pKa = 4.44VAGAPGMVQDD139 pKa = 4.69SIWRR143 pKa = 11.84TRR145 pKa = 11.84TVSVGRR151 pKa = 11.84GRR153 pKa = 11.84GNSTQNAIVDD163 pKa = 3.73PTKK166 pKa = 10.36YY167 pKa = 10.55GHH169 pKa = 5.44YY170 pKa = 8.98TSTAGEE176 pKa = 3.97PRR178 pKa = 11.84FYY180 pKa = 11.2SPGSIRR186 pKa = 11.84ALVDD190 pKa = 3.12GRR192 pKa = 11.84VGQAGTMVLYY202 pKa = 8.63CTYY205 pKa = 10.65RR206 pKa = 11.84CTLHH210 pKa = 5.8VQARR214 pKa = 11.84QDD216 pKa = 3.75DD217 pKa = 4.42AQLAPAPVIRR227 pKa = 11.84NVFPWVWPDD236 pKa = 3.26DD237 pKa = 3.53TDD239 pKa = 4.54YY240 pKa = 11.61AITDD244 pKa = 4.15PGPNGVVIRR253 pKa = 11.84WDD255 pKa = 3.46DD256 pKa = 3.46SFPEE260 pKa = 4.31YY261 pKa = 10.42EE262 pKa = 4.82RR263 pKa = 11.84PTTDD267 pKa = 2.37TVFQVPVPVVCSAKK281 pKa = 9.18GTNAGGRR288 pKa = 11.84VICPYY293 pKa = 10.37ILYY296 pKa = 10.07QPSSDD301 pKa = 3.51QFYY304 pKa = 8.11MTDD307 pKa = 4.03DD308 pKa = 3.94LDD310 pKa = 4.14GAAVLQATSPSAASVFQIMPAGMTFTVVYY339 pKa = 9.9TEE341 pKa = 4.57GEE343 pKa = 4.16VTRR346 pKa = 11.84GVRR349 pKa = 11.84EE350 pKa = 3.99LPGGGLTNRR359 pKa = 11.84PPAPRR364 pKa = 11.84GIRR367 pKa = 11.84RR368 pKa = 11.84KK369 pKa = 10.24AYY371 pKa = 9.79
MM1 pKa = 7.35TARR4 pKa = 11.84CVLAMPRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GTATNGAGYY22 pKa = 10.78APVSLGTGVRR32 pKa = 11.84ANRR35 pKa = 11.84RR36 pKa = 11.84TPPMQLEE43 pKa = 4.23GTDD46 pKa = 3.62RR47 pKa = 11.84IGVINWDD54 pKa = 3.47GALDD58 pKa = 3.78TGHH61 pKa = 6.92VLFDD65 pKa = 4.23LLVEE69 pKa = 4.2PGIFKK74 pKa = 10.6SLRR77 pKa = 11.84DD78 pKa = 3.52VAKK81 pKa = 10.41ARR83 pKa = 11.84QYY85 pKa = 9.87IQWHH89 pKa = 5.12ALEE92 pKa = 5.26FKK94 pKa = 10.25FSCGTNTAKK103 pKa = 10.76DD104 pKa = 3.84GSYY107 pKa = 10.38VAAFIADD114 pKa = 4.17PDD116 pKa = 4.58DD117 pKa = 5.0KK118 pKa = 11.76NPDD121 pKa = 3.76DD122 pKa = 3.69PTLAVEE128 pKa = 4.81EE129 pKa = 4.44VAGAPGMVQDD139 pKa = 4.69SIWRR143 pKa = 11.84TRR145 pKa = 11.84TVSVGRR151 pKa = 11.84GRR153 pKa = 11.84GNSTQNAIVDD163 pKa = 3.73PTKK166 pKa = 10.36YY167 pKa = 10.55GHH169 pKa = 5.44YY170 pKa = 8.98TSTAGEE176 pKa = 3.97PRR178 pKa = 11.84FYY180 pKa = 11.2SPGSIRR186 pKa = 11.84ALVDD190 pKa = 3.12GRR192 pKa = 11.84VGQAGTMVLYY202 pKa = 8.63CTYY205 pKa = 10.65RR206 pKa = 11.84CTLHH210 pKa = 5.8VQARR214 pKa = 11.84QDD216 pKa = 3.75DD217 pKa = 4.42AQLAPAPVIRR227 pKa = 11.84NVFPWVWPDD236 pKa = 3.26DD237 pKa = 3.53TDD239 pKa = 4.54YY240 pKa = 11.61AITDD244 pKa = 4.15PGPNGVVIRR253 pKa = 11.84WDD255 pKa = 3.46DD256 pKa = 3.46SFPEE260 pKa = 4.31YY261 pKa = 10.42EE262 pKa = 4.82RR263 pKa = 11.84PTTDD267 pKa = 2.37TVFQVPVPVVCSAKK281 pKa = 9.18GTNAGGRR288 pKa = 11.84VICPYY293 pKa = 10.37ILYY296 pKa = 10.07QPSSDD301 pKa = 3.51QFYY304 pKa = 8.11MTDD307 pKa = 4.03DD308 pKa = 3.94LDD310 pKa = 4.14GAAVLQATSPSAASVFQIMPAGMTFTVVYY339 pKa = 9.9TEE341 pKa = 4.57GEE343 pKa = 4.16VTRR346 pKa = 11.84GVRR349 pKa = 11.84EE350 pKa = 3.99LPGGGLTNRR359 pKa = 11.84PPAPRR364 pKa = 11.84GIRR367 pKa = 11.84RR368 pKa = 11.84KK369 pKa = 10.24AYY371 pKa = 9.79
Molecular weight: 40.26 kDa
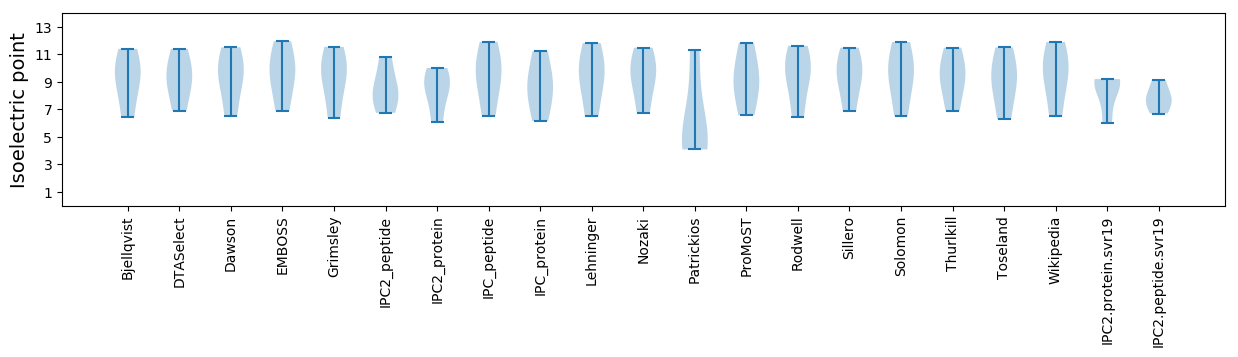
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGH8|A0A1L3KGH8_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 7 OX=1923391 PE=4 SV=1
MM1 pKa = 7.4ATTAQAGGSKK11 pKa = 10.0APKK14 pKa = 9.43GARR17 pKa = 11.84PVQTGNSANGVSGSNASMKK36 pKa = 10.38KK37 pKa = 9.87ILDD40 pKa = 3.99ALPGVSPKK48 pKa = 10.06FVRR51 pKa = 11.84RR52 pKa = 11.84VLRR55 pKa = 11.84GRR57 pKa = 11.84RR58 pKa = 11.84LGITATVLAVYY69 pKa = 10.1NAIQRR74 pKa = 11.84ADD76 pKa = 3.92PEE78 pKa = 4.71DD79 pKa = 5.1PICSSRR85 pKa = 11.84SHH87 pKa = 5.17TNKK90 pKa = 9.39WLNAHH95 pKa = 6.57LNGVASQRR103 pKa = 11.84QAKK106 pKa = 8.3TKK108 pKa = 10.46GRR110 pKa = 11.84TEE112 pKa = 3.91PLIKK116 pKa = 10.16QAADD120 pKa = 3.26EE121 pKa = 4.46LPNNKK126 pKa = 8.32RR127 pKa = 11.84TTGKK131 pKa = 10.6AEE133 pKa = 4.15PKK135 pKa = 9.99QGNAKK140 pKa = 9.69SRR142 pKa = 11.84RR143 pKa = 11.84AEE145 pKa = 3.92RR146 pKa = 11.84RR147 pKa = 11.84AHH149 pKa = 5.84LQPLVRR155 pKa = 11.84GADD158 pKa = 3.3AKK160 pKa = 10.83LAAAKK165 pKa = 9.82HH166 pKa = 5.65RR167 pKa = 11.84SAGGPAMGMSVAWQAEE183 pKa = 4.45ANKK186 pKa = 10.6APLPVTKK193 pKa = 10.24RR194 pKa = 11.84RR195 pKa = 11.84PKK197 pKa = 9.87EE198 pKa = 3.66SRR200 pKa = 11.84IKK202 pKa = 10.42YY203 pKa = 9.73LIRR206 pKa = 11.84SYY208 pKa = 10.73EE209 pKa = 3.88AVIARR214 pKa = 11.84GDD216 pKa = 3.61KK217 pKa = 10.41LHH219 pKa = 5.67QQGKK223 pKa = 9.0IVPEE227 pKa = 4.0NLHH230 pKa = 5.98LQADD234 pKa = 3.84KK235 pKa = 11.48LEE237 pKa = 4.21NLLRR241 pKa = 11.84EE242 pKa = 4.14EE243 pKa = 4.18RR244 pKa = 11.84EE245 pKa = 4.01KK246 pKa = 11.27VRR248 pKa = 11.84GHH250 pKa = 5.95NKK252 pKa = 9.34VNPVRR257 pKa = 11.84LNAKK261 pKa = 9.35KK262 pKa = 10.09RR263 pKa = 11.84RR264 pKa = 11.84SGLRR268 pKa = 11.84RR269 pKa = 11.84QNPEE273 pKa = 3.74LEE275 pKa = 4.16AVRR278 pKa = 11.84SKK280 pKa = 10.74RR281 pKa = 11.84AAAVPMNLNKK291 pKa = 10.52LLGQTQGPLSSVLGNAGLAKK311 pKa = 9.45TVEE314 pKa = 4.61SILPEE319 pKa = 4.26NGLTVTIKK327 pKa = 10.47PSSQRR332 pKa = 11.84EE333 pKa = 3.81RR334 pKa = 11.84KK335 pKa = 8.83NARR338 pKa = 11.84KK339 pKa = 9.17AQVQRR344 pKa = 11.84KK345 pKa = 7.87
MM1 pKa = 7.4ATTAQAGGSKK11 pKa = 10.0APKK14 pKa = 9.43GARR17 pKa = 11.84PVQTGNSANGVSGSNASMKK36 pKa = 10.38KK37 pKa = 9.87ILDD40 pKa = 3.99ALPGVSPKK48 pKa = 10.06FVRR51 pKa = 11.84RR52 pKa = 11.84VLRR55 pKa = 11.84GRR57 pKa = 11.84RR58 pKa = 11.84LGITATVLAVYY69 pKa = 10.1NAIQRR74 pKa = 11.84ADD76 pKa = 3.92PEE78 pKa = 4.71DD79 pKa = 5.1PICSSRR85 pKa = 11.84SHH87 pKa = 5.17TNKK90 pKa = 9.39WLNAHH95 pKa = 6.57LNGVASQRR103 pKa = 11.84QAKK106 pKa = 8.3TKK108 pKa = 10.46GRR110 pKa = 11.84TEE112 pKa = 3.91PLIKK116 pKa = 10.16QAADD120 pKa = 3.26EE121 pKa = 4.46LPNNKK126 pKa = 8.32RR127 pKa = 11.84TTGKK131 pKa = 10.6AEE133 pKa = 4.15PKK135 pKa = 9.99QGNAKK140 pKa = 9.69SRR142 pKa = 11.84RR143 pKa = 11.84AEE145 pKa = 3.92RR146 pKa = 11.84RR147 pKa = 11.84AHH149 pKa = 5.84LQPLVRR155 pKa = 11.84GADD158 pKa = 3.3AKK160 pKa = 10.83LAAAKK165 pKa = 9.82HH166 pKa = 5.65RR167 pKa = 11.84SAGGPAMGMSVAWQAEE183 pKa = 4.45ANKK186 pKa = 10.6APLPVTKK193 pKa = 10.24RR194 pKa = 11.84RR195 pKa = 11.84PKK197 pKa = 9.87EE198 pKa = 3.66SRR200 pKa = 11.84IKK202 pKa = 10.42YY203 pKa = 9.73LIRR206 pKa = 11.84SYY208 pKa = 10.73EE209 pKa = 3.88AVIARR214 pKa = 11.84GDD216 pKa = 3.61KK217 pKa = 10.41LHH219 pKa = 5.67QQGKK223 pKa = 9.0IVPEE227 pKa = 4.0NLHH230 pKa = 5.98LQADD234 pKa = 3.84KK235 pKa = 11.48LEE237 pKa = 4.21NLLRR241 pKa = 11.84EE242 pKa = 4.14EE243 pKa = 4.18RR244 pKa = 11.84EE245 pKa = 4.01KK246 pKa = 11.27VRR248 pKa = 11.84GHH250 pKa = 5.95NKK252 pKa = 9.34VNPVRR257 pKa = 11.84LNAKK261 pKa = 9.35KK262 pKa = 10.09RR263 pKa = 11.84RR264 pKa = 11.84SGLRR268 pKa = 11.84RR269 pKa = 11.84QNPEE273 pKa = 3.74LEE275 pKa = 4.16AVRR278 pKa = 11.84SKK280 pKa = 10.74RR281 pKa = 11.84AAAVPMNLNKK291 pKa = 10.52LLGQTQGPLSSVLGNAGLAKK311 pKa = 9.45TVEE314 pKa = 4.61SILPEE319 pKa = 4.26NGLTVTIKK327 pKa = 10.47PSSQRR332 pKa = 11.84EE333 pKa = 3.81RR334 pKa = 11.84KK335 pKa = 8.83NARR338 pKa = 11.84KK339 pKa = 9.17AQVQRR344 pKa = 11.84KK345 pKa = 7.87
Molecular weight: 37.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1722 |
345 |
527 |
430.5 |
47.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.266 ± 0.826 | 1.626 ± 0.373 |
4.413 ± 0.891 | 5.285 ± 0.704 |
2.381 ± 0.781 | 7.491 ± 0.871 |
2.439 ± 0.476 | 3.194 ± 0.442 |
5.11 ± 1.072 | 7.085 ± 1.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.469 | 4.181 ± 0.485 |
6.214 ± 0.658 | 4.297 ± 0.605 |
9.001 ± 0.339 | 6.33 ± 0.493 |
5.691 ± 0.603 | 7.607 ± 0.605 |
1.568 ± 0.246 | 2.613 ± 0.436 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
