
Heterobasidion partitivirus 12
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
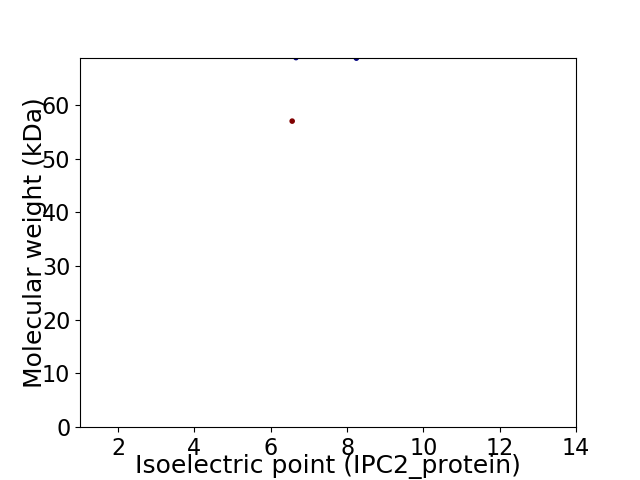
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
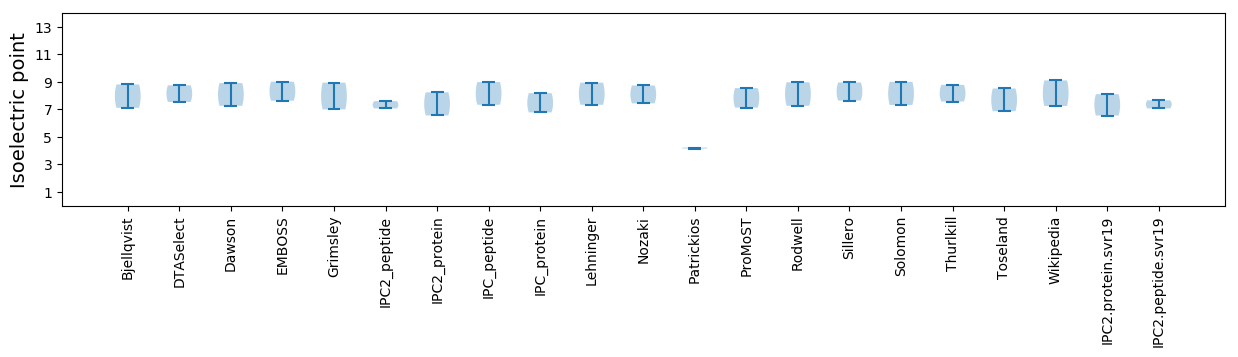
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8PR73|W8PR73_9VIRU Capsid protein OS=Heterobasidion partitivirus 12 OX=1469905 PE=4 SV=1
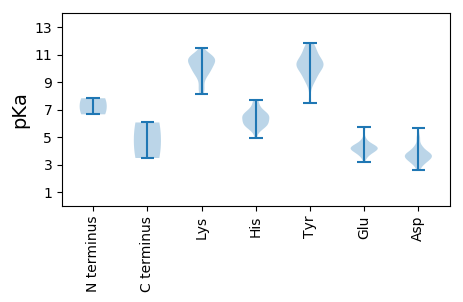
MM1 pKa = 7.82SSLPDD6 pKa = 3.58FASMNEE12 pKa = 4.04SEE14 pKa = 4.06RR15 pKa = 11.84LKK17 pKa = 11.04YY18 pKa = 10.54FEE20 pKa = 4.48EE21 pKa = 4.17QQKK24 pKa = 9.61KK25 pKa = 9.44QKK27 pKa = 10.85DD28 pKa = 3.68LVDD31 pKa = 3.55QSIKK35 pKa = 10.85AKK37 pKa = 8.52QTMSAMRR44 pKa = 11.84VRR46 pKa = 11.84PVFQQAIDD54 pKa = 3.93GAKK57 pKa = 9.96SASKK61 pKa = 10.64SSAVPTKK68 pKa = 10.44AVPPPPFSGTSASANNMLRR87 pKa = 11.84LAFGEE92 pKa = 4.62SPFRR96 pKa = 11.84PKK98 pKa = 10.41QRR100 pKa = 11.84FGQNGFIPDD109 pKa = 3.69SQFLFTLLSIMDD121 pKa = 3.79MKK123 pKa = 9.28MTSTRR128 pKa = 11.84KK129 pKa = 9.58FIEE132 pKa = 4.55GAEE135 pKa = 4.24FWSPLISQYY144 pKa = 11.59YY145 pKa = 7.47MAVLVFVQIFRR156 pKa = 11.84AMHH159 pKa = 5.53EE160 pKa = 4.35AGLLSGEE167 pKa = 4.17AADD170 pKa = 5.64VYY172 pKa = 11.58DD173 pKa = 4.91LFCGHH178 pKa = 6.36SPAISLASLPVPGPFLNLLAQYY200 pKa = 10.05AVHH203 pKa = 6.68IPHH206 pKa = 7.04LVDD209 pKa = 3.84MDD211 pKa = 5.15NICPMIPNNLDD222 pKa = 3.24TTNTNFYY229 pKa = 10.06KK230 pKa = 10.82FQGLCSNLLGRR241 pKa = 11.84LPNVPYY247 pKa = 10.88LLDD250 pKa = 3.8QIALLRR256 pKa = 11.84DD257 pKa = 3.65NLVAAPVSIDD267 pKa = 3.18VANQGRR273 pKa = 11.84VLHH276 pKa = 5.81RR277 pKa = 11.84TMFGQALFAINAAPGNAAQWNTRR300 pKa = 11.84VPYY303 pKa = 10.2GAAVPDD309 pKa = 3.5NAKK312 pKa = 10.73YY313 pKa = 10.28ISCDD317 pKa = 3.05PASRR321 pKa = 11.84HH322 pKa = 5.69PFFLNRR328 pKa = 11.84GLISQMTDD336 pKa = 2.74YY337 pKa = 11.39AALINVNTPAAHH349 pKa = 6.12NAAAITPSWTQFLGLDD365 pKa = 3.44NMPFFLQVVRR375 pKa = 11.84IMSWYY380 pKa = 9.16SKK382 pKa = 9.33FWQGSTDD389 pKa = 3.58LASFSPNGHH398 pKa = 5.99TSGQNTFTLVNKK410 pKa = 8.75PANLAAITDD419 pKa = 3.51ARR421 pKa = 11.84FYY423 pKa = 11.53SLDD426 pKa = 3.44LKK428 pKa = 10.76MKK430 pKa = 10.13GVARR434 pKa = 11.84NPLAPEE440 pKa = 4.08SDD442 pKa = 3.73SFDD445 pKa = 3.71AATGPINLSWTPADD459 pKa = 3.6MYY461 pKa = 10.98GGKK464 pKa = 9.99LDD466 pKa = 3.75TTAITATEE474 pKa = 4.36ANLSTHH480 pKa = 5.8RR481 pKa = 11.84QGGPFFTEE489 pKa = 4.21SIVDD493 pKa = 3.22ITGEE497 pKa = 4.02INPTSAYY504 pKa = 10.54AGILNQYY511 pKa = 8.75YY512 pKa = 9.59YY513 pKa = 11.47SSVALKK519 pKa = 10.59NN520 pKa = 3.48
MM1 pKa = 7.82SSLPDD6 pKa = 3.58FASMNEE12 pKa = 4.04SEE14 pKa = 4.06RR15 pKa = 11.84LKK17 pKa = 11.04YY18 pKa = 10.54FEE20 pKa = 4.48EE21 pKa = 4.17QQKK24 pKa = 9.61KK25 pKa = 9.44QKK27 pKa = 10.85DD28 pKa = 3.68LVDD31 pKa = 3.55QSIKK35 pKa = 10.85AKK37 pKa = 8.52QTMSAMRR44 pKa = 11.84VRR46 pKa = 11.84PVFQQAIDD54 pKa = 3.93GAKK57 pKa = 9.96SASKK61 pKa = 10.64SSAVPTKK68 pKa = 10.44AVPPPPFSGTSASANNMLRR87 pKa = 11.84LAFGEE92 pKa = 4.62SPFRR96 pKa = 11.84PKK98 pKa = 10.41QRR100 pKa = 11.84FGQNGFIPDD109 pKa = 3.69SQFLFTLLSIMDD121 pKa = 3.79MKK123 pKa = 9.28MTSTRR128 pKa = 11.84KK129 pKa = 9.58FIEE132 pKa = 4.55GAEE135 pKa = 4.24FWSPLISQYY144 pKa = 11.59YY145 pKa = 7.47MAVLVFVQIFRR156 pKa = 11.84AMHH159 pKa = 5.53EE160 pKa = 4.35AGLLSGEE167 pKa = 4.17AADD170 pKa = 5.64VYY172 pKa = 11.58DD173 pKa = 4.91LFCGHH178 pKa = 6.36SPAISLASLPVPGPFLNLLAQYY200 pKa = 10.05AVHH203 pKa = 6.68IPHH206 pKa = 7.04LVDD209 pKa = 3.84MDD211 pKa = 5.15NICPMIPNNLDD222 pKa = 3.24TTNTNFYY229 pKa = 10.06KK230 pKa = 10.82FQGLCSNLLGRR241 pKa = 11.84LPNVPYY247 pKa = 10.88LLDD250 pKa = 3.8QIALLRR256 pKa = 11.84DD257 pKa = 3.65NLVAAPVSIDD267 pKa = 3.18VANQGRR273 pKa = 11.84VLHH276 pKa = 5.81RR277 pKa = 11.84TMFGQALFAINAAPGNAAQWNTRR300 pKa = 11.84VPYY303 pKa = 10.2GAAVPDD309 pKa = 3.5NAKK312 pKa = 10.73YY313 pKa = 10.28ISCDD317 pKa = 3.05PASRR321 pKa = 11.84HH322 pKa = 5.69PFFLNRR328 pKa = 11.84GLISQMTDD336 pKa = 2.74YY337 pKa = 11.39AALINVNTPAAHH349 pKa = 6.12NAAAITPSWTQFLGLDD365 pKa = 3.44NMPFFLQVVRR375 pKa = 11.84IMSWYY380 pKa = 9.16SKK382 pKa = 9.33FWQGSTDD389 pKa = 3.58LASFSPNGHH398 pKa = 5.99TSGQNTFTLVNKK410 pKa = 8.75PANLAAITDD419 pKa = 3.51ARR421 pKa = 11.84FYY423 pKa = 11.53SLDD426 pKa = 3.44LKK428 pKa = 10.76MKK430 pKa = 10.13GVARR434 pKa = 11.84NPLAPEE440 pKa = 4.08SDD442 pKa = 3.73SFDD445 pKa = 3.71AATGPINLSWTPADD459 pKa = 3.6MYY461 pKa = 10.98GGKK464 pKa = 9.99LDD466 pKa = 3.75TTAITATEE474 pKa = 4.36ANLSTHH480 pKa = 5.8RR481 pKa = 11.84QGGPFFTEE489 pKa = 4.21SIVDD493 pKa = 3.22ITGEE497 pKa = 4.02INPTSAYY504 pKa = 10.54AGILNQYY511 pKa = 8.75YY512 pKa = 9.59YY513 pKa = 11.47SSVALKK519 pKa = 10.59NN520 pKa = 3.48
Molecular weight: 56.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8PR73|W8PR73_9VIRU Capsid protein OS=Heterobasidion partitivirus 12 OX=1469905 PE=4 SV=1
MM1 pKa = 6.65QTLLSAVYY9 pKa = 10.07SIRR12 pKa = 11.84DD13 pKa = 3.74AIFGKK18 pKa = 8.31TLSEE22 pKa = 3.74PHH24 pKa = 5.94GLNVNFHH31 pKa = 6.78FDD33 pKa = 3.41GYY35 pKa = 9.56VTDD38 pKa = 4.91EE39 pKa = 4.82IKK41 pKa = 10.17TSIPPRR47 pKa = 11.84VEE49 pKa = 3.19MWYY52 pKa = 10.69SDD54 pKa = 3.59YY55 pKa = 11.48QKK57 pKa = 10.78FLNPIIRR64 pKa = 11.84ANFTGAEE71 pKa = 3.86ADD73 pKa = 3.96KK74 pKa = 10.79IINGYY79 pKa = 7.67HH80 pKa = 6.3HH81 pKa = 7.62PVATIPFMVEE91 pKa = 3.51NLKK94 pKa = 10.99KK95 pKa = 10.65GDD97 pKa = 4.09LSDD100 pKa = 4.26HH101 pKa = 6.45PVPHH105 pKa = 7.3DD106 pKa = 3.44EE107 pKa = 4.27HH108 pKa = 7.69YY109 pKa = 11.18LRR111 pKa = 11.84ARR113 pKa = 11.84KK114 pKa = 8.14MAADD118 pKa = 4.28AFRR121 pKa = 11.84PPRR124 pKa = 11.84PIRR127 pKa = 11.84PVHH130 pKa = 5.54FADD133 pKa = 4.29LRR135 pKa = 11.84FYY137 pKa = 10.34NWNWHH142 pKa = 6.18PNVEE146 pKa = 4.03EE147 pKa = 4.33PYY149 pKa = 9.99YY150 pKa = 10.35SNKK153 pKa = 9.61RR154 pKa = 11.84AQEE157 pKa = 4.14YY158 pKa = 9.3VQAAHH163 pKa = 6.44TLGLTPDD170 pKa = 3.18ARR172 pKa = 11.84MSFGNLRR179 pKa = 11.84DD180 pKa = 3.71YY181 pKa = 11.82VFMDD185 pKa = 3.24TRR187 pKa = 11.84HH188 pKa = 5.91YY189 pKa = 10.6LHH191 pKa = 6.45QIKK194 pKa = 10.29RR195 pKa = 11.84NEE197 pKa = 3.67ISNPKK202 pKa = 8.3TLWPLMKK209 pKa = 9.9IHH211 pKa = 6.61VKK213 pKa = 9.84PALTGTDD220 pKa = 3.31EE221 pKa = 3.97QKK223 pKa = 10.35IRR225 pKa = 11.84VIYY228 pKa = 9.84GVSKK232 pKa = 10.55RR233 pKa = 11.84HH234 pKa = 5.93VLPQCMFFWPLFRR247 pKa = 11.84HH248 pKa = 5.89YY249 pKa = 10.4IEE251 pKa = 5.74NDD253 pKa = 3.07TDD255 pKa = 3.84PLLWGFEE262 pKa = 4.32TILGGMMKK270 pKa = 10.3LNSLMHH276 pKa = 6.73RR277 pKa = 11.84LYY279 pKa = 10.19FQTFVTVDD287 pKa = 2.64WSGFDD292 pKa = 3.55LRR294 pKa = 11.84SLFSIQRR301 pKa = 11.84EE302 pKa = 4.03IFDD305 pKa = 3.3DD306 pKa = 2.94WRR308 pKa = 11.84TYY310 pKa = 10.99FDD312 pKa = 4.36FSNGYY317 pKa = 9.29IPTVWYY323 pKa = 8.88PNSTADD329 pKa = 4.25PIQLEE334 pKa = 4.52RR335 pKa = 11.84LWNWQRR341 pKa = 11.84DD342 pKa = 3.54ACFNMPFVMPDD353 pKa = 2.68KK354 pKa = 10.69SVYY357 pKa = 9.76RR358 pKa = 11.84RR359 pKa = 11.84LFRR362 pKa = 11.84AIPSGLFVTQFLDD375 pKa = 3.21SHH377 pKa = 6.7YY378 pKa = 10.74NYY380 pKa = 10.34IMLLTILSAMGFDD393 pKa = 3.29ITIEE397 pKa = 4.13RR398 pKa = 11.84IRR400 pKa = 11.84ILVQGDD406 pKa = 3.91DD407 pKa = 3.73SLKK410 pKa = 10.81NLIFFIPANQHH421 pKa = 6.58DD422 pKa = 4.28NFKK425 pKa = 11.45AEE427 pKa = 4.29FQRR430 pKa = 11.84LATYY434 pKa = 10.81YY435 pKa = 9.15FDD437 pKa = 3.98HH438 pKa = 6.68VARR441 pKa = 11.84PEE443 pKa = 3.64KK444 pKa = 10.28TEE446 pKa = 4.35IYY448 pKa = 10.37NSPQGVTVLGYY459 pKa = 9.26TNNNGFPTRR468 pKa = 11.84DD469 pKa = 3.91PIKK472 pKa = 10.93LMAQLYY478 pKa = 9.53HH479 pKa = 6.54PRR481 pKa = 11.84QVGEE485 pKa = 3.82RR486 pKa = 11.84WKK488 pKa = 10.68SVLMAKK494 pKa = 10.26CAGFAYY500 pKa = 10.12ASAYY504 pKa = 10.34NYY506 pKa = 9.7PKK508 pKa = 9.6VTATLKK514 pKa = 8.79TVYY517 pKa = 10.28YY518 pKa = 10.16KK519 pKa = 10.9LAAKK523 pKa = 10.0GFSPAALRR531 pKa = 11.84TQRR534 pKa = 11.84DD535 pKa = 3.05IVLFGEE541 pKa = 4.92AKK543 pKa = 9.82FQVPTDD549 pKa = 3.74HH550 pKa = 7.07FPTAEE555 pKa = 3.8EE556 pKa = 4.32VQRR559 pKa = 11.84HH560 pKa = 4.9LRR562 pKa = 11.84VPYY565 pKa = 10.16KK566 pKa = 9.9RR567 pKa = 11.84TEE569 pKa = 4.09EE570 pKa = 4.17DD571 pKa = 3.45SEE573 pKa = 5.6SYY575 pKa = 10.98FPMKK579 pKa = 10.46HH580 pKa = 6.05FLDD583 pKa = 4.86FAA585 pKa = 6.06
MM1 pKa = 6.65QTLLSAVYY9 pKa = 10.07SIRR12 pKa = 11.84DD13 pKa = 3.74AIFGKK18 pKa = 8.31TLSEE22 pKa = 3.74PHH24 pKa = 5.94GLNVNFHH31 pKa = 6.78FDD33 pKa = 3.41GYY35 pKa = 9.56VTDD38 pKa = 4.91EE39 pKa = 4.82IKK41 pKa = 10.17TSIPPRR47 pKa = 11.84VEE49 pKa = 3.19MWYY52 pKa = 10.69SDD54 pKa = 3.59YY55 pKa = 11.48QKK57 pKa = 10.78FLNPIIRR64 pKa = 11.84ANFTGAEE71 pKa = 3.86ADD73 pKa = 3.96KK74 pKa = 10.79IINGYY79 pKa = 7.67HH80 pKa = 6.3HH81 pKa = 7.62PVATIPFMVEE91 pKa = 3.51NLKK94 pKa = 10.99KK95 pKa = 10.65GDD97 pKa = 4.09LSDD100 pKa = 4.26HH101 pKa = 6.45PVPHH105 pKa = 7.3DD106 pKa = 3.44EE107 pKa = 4.27HH108 pKa = 7.69YY109 pKa = 11.18LRR111 pKa = 11.84ARR113 pKa = 11.84KK114 pKa = 8.14MAADD118 pKa = 4.28AFRR121 pKa = 11.84PPRR124 pKa = 11.84PIRR127 pKa = 11.84PVHH130 pKa = 5.54FADD133 pKa = 4.29LRR135 pKa = 11.84FYY137 pKa = 10.34NWNWHH142 pKa = 6.18PNVEE146 pKa = 4.03EE147 pKa = 4.33PYY149 pKa = 9.99YY150 pKa = 10.35SNKK153 pKa = 9.61RR154 pKa = 11.84AQEE157 pKa = 4.14YY158 pKa = 9.3VQAAHH163 pKa = 6.44TLGLTPDD170 pKa = 3.18ARR172 pKa = 11.84MSFGNLRR179 pKa = 11.84DD180 pKa = 3.71YY181 pKa = 11.82VFMDD185 pKa = 3.24TRR187 pKa = 11.84HH188 pKa = 5.91YY189 pKa = 10.6LHH191 pKa = 6.45QIKK194 pKa = 10.29RR195 pKa = 11.84NEE197 pKa = 3.67ISNPKK202 pKa = 8.3TLWPLMKK209 pKa = 9.9IHH211 pKa = 6.61VKK213 pKa = 9.84PALTGTDD220 pKa = 3.31EE221 pKa = 3.97QKK223 pKa = 10.35IRR225 pKa = 11.84VIYY228 pKa = 9.84GVSKK232 pKa = 10.55RR233 pKa = 11.84HH234 pKa = 5.93VLPQCMFFWPLFRR247 pKa = 11.84HH248 pKa = 5.89YY249 pKa = 10.4IEE251 pKa = 5.74NDD253 pKa = 3.07TDD255 pKa = 3.84PLLWGFEE262 pKa = 4.32TILGGMMKK270 pKa = 10.3LNSLMHH276 pKa = 6.73RR277 pKa = 11.84LYY279 pKa = 10.19FQTFVTVDD287 pKa = 2.64WSGFDD292 pKa = 3.55LRR294 pKa = 11.84SLFSIQRR301 pKa = 11.84EE302 pKa = 4.03IFDD305 pKa = 3.3DD306 pKa = 2.94WRR308 pKa = 11.84TYY310 pKa = 10.99FDD312 pKa = 4.36FSNGYY317 pKa = 9.29IPTVWYY323 pKa = 8.88PNSTADD329 pKa = 4.25PIQLEE334 pKa = 4.52RR335 pKa = 11.84LWNWQRR341 pKa = 11.84DD342 pKa = 3.54ACFNMPFVMPDD353 pKa = 2.68KK354 pKa = 10.69SVYY357 pKa = 9.76RR358 pKa = 11.84RR359 pKa = 11.84LFRR362 pKa = 11.84AIPSGLFVTQFLDD375 pKa = 3.21SHH377 pKa = 6.7YY378 pKa = 10.74NYY380 pKa = 10.34IMLLTILSAMGFDD393 pKa = 3.29ITIEE397 pKa = 4.13RR398 pKa = 11.84IRR400 pKa = 11.84ILVQGDD406 pKa = 3.91DD407 pKa = 3.73SLKK410 pKa = 10.81NLIFFIPANQHH421 pKa = 6.58DD422 pKa = 4.28NFKK425 pKa = 11.45AEE427 pKa = 4.29FQRR430 pKa = 11.84LATYY434 pKa = 10.81YY435 pKa = 9.15FDD437 pKa = 3.98HH438 pKa = 6.68VARR441 pKa = 11.84PEE443 pKa = 3.64KK444 pKa = 10.28TEE446 pKa = 4.35IYY448 pKa = 10.37NSPQGVTVLGYY459 pKa = 9.26TNNNGFPTRR468 pKa = 11.84DD469 pKa = 3.91PIKK472 pKa = 10.93LMAQLYY478 pKa = 9.53HH479 pKa = 6.54PRR481 pKa = 11.84QVGEE485 pKa = 3.82RR486 pKa = 11.84WKK488 pKa = 10.68SVLMAKK494 pKa = 10.26CAGFAYY500 pKa = 10.12ASAYY504 pKa = 10.34NYY506 pKa = 9.7PKK508 pKa = 9.6VTATLKK514 pKa = 8.79TVYY517 pKa = 10.28YY518 pKa = 10.16KK519 pKa = 10.9LAAKK523 pKa = 10.0GFSPAALRR531 pKa = 11.84TQRR534 pKa = 11.84DD535 pKa = 3.05IVLFGEE541 pKa = 4.92AKK543 pKa = 9.82FQVPTDD549 pKa = 3.74HH550 pKa = 7.07FPTAEE555 pKa = 3.8EE556 pKa = 4.32VQRR559 pKa = 11.84HH560 pKa = 4.9LRR562 pKa = 11.84VPYY565 pKa = 10.16KK566 pKa = 9.9RR567 pKa = 11.84TEE569 pKa = 4.09EE570 pKa = 4.17DD571 pKa = 3.45SEE573 pKa = 5.6SYY575 pKa = 10.98FPMKK579 pKa = 10.46HH580 pKa = 6.05FLDD583 pKa = 4.86FAA585 pKa = 6.06
Molecular weight: 68.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1105 |
520 |
585 |
552.5 |
62.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.688 ± 1.65 | 0.633 ± 0.091 |
5.52 ± 0.348 | 3.529 ± 0.689 |
6.697 ± 0.363 | 4.887 ± 0.462 |
2.896 ± 0.779 | 5.339 ± 0.227 |
4.434 ± 0.393 | 8.688 ± 0.363 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.258 ± 0.136 | 5.52 ± 0.552 |
6.878 ± 0.159 | 4.163 ± 0.431 |
5.158 ± 1.006 | 6.425 ± 1.362 |
5.792 ± 0.015 | 5.339 ± 0.227 |
1.629 ± 0.318 | 4.525 ± 0.84 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
