
Punctularia strigosozonata (strain HHB-11173) (White-rot fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Corticiales; Punctulariaceae; Punctularia; Punctularia strigosozonata
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
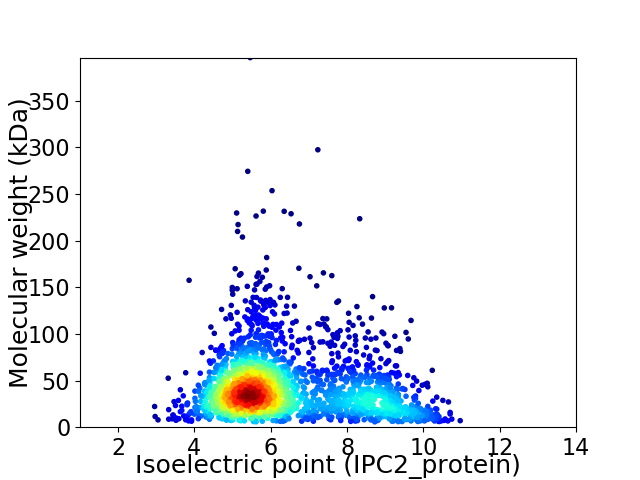
Virtual 2D-PAGE plot for 2093 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7RZT3|R7RZT3_PUNST Uncharacterized protein OS=Punctularia strigosozonata (strain HHB-11173) OX=741275 GN=PUNSTDRAFT_146953 PE=4 SV=1
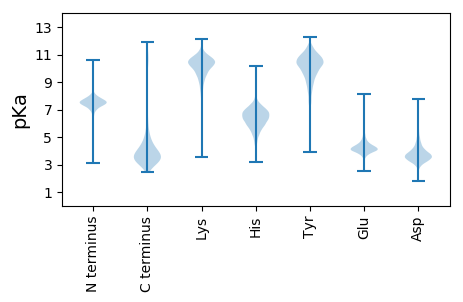
MM1 pKa = 7.8LSPLIPLTLTVAIATLSHH19 pKa = 7.14ADD21 pKa = 3.4TVVIPTSSFDD31 pKa = 3.43SYY33 pKa = 12.14ADD35 pKa = 4.19LEE37 pKa = 4.85TYY39 pKa = 9.33WNYY42 pKa = 10.98LYY44 pKa = 10.25PWGSDD49 pKa = 3.3HH50 pKa = 6.98NGSARR55 pKa = 11.84MVGNSTDD62 pKa = 3.17HH63 pKa = 5.96SHH65 pKa = 6.95ISVANGVLTLTATPTSGQGTSSEE88 pKa = 4.52SPHH91 pKa = 5.72LTIDD95 pKa = 3.71YY96 pKa = 10.5FSGTIYY102 pKa = 10.86AKK104 pKa = 10.74DD105 pKa = 4.65DD106 pKa = 3.41ITVDD110 pKa = 3.23GTDD113 pKa = 3.1VVGYY117 pKa = 9.1VVEE120 pKa = 4.71GEE122 pKa = 4.9FIAPTAVGTWPAFWLTAVNSWPPEE146 pKa = 3.67ADD148 pKa = 2.63IGEE151 pKa = 4.35WKK153 pKa = 9.01GTQQTWFNTFNTSSEE168 pKa = 4.27VKK170 pKa = 9.03STIVAWPDD178 pKa = 3.04DD179 pKa = 4.25GEE181 pKa = 4.3FHH183 pKa = 6.73SVKK186 pKa = 10.51AVLSSIEE193 pKa = 4.15GDD195 pKa = 3.63SSDD198 pKa = 3.67VTIDD202 pKa = 3.63YY203 pKa = 11.21YY204 pKa = 11.46LDD206 pKa = 3.42NVLQATHH213 pKa = 6.19VGADD217 pKa = 3.57FLGAPLYY224 pKa = 10.65LIIDD228 pKa = 4.24LQMEE232 pKa = 5.0GSSGSPGPTGTTTYY246 pKa = 10.51QIRR249 pKa = 11.84NVQVTKK255 pKa = 10.12VTSS258 pKa = 3.3
MM1 pKa = 7.8LSPLIPLTLTVAIATLSHH19 pKa = 7.14ADD21 pKa = 3.4TVVIPTSSFDD31 pKa = 3.43SYY33 pKa = 12.14ADD35 pKa = 4.19LEE37 pKa = 4.85TYY39 pKa = 9.33WNYY42 pKa = 10.98LYY44 pKa = 10.25PWGSDD49 pKa = 3.3HH50 pKa = 6.98NGSARR55 pKa = 11.84MVGNSTDD62 pKa = 3.17HH63 pKa = 5.96SHH65 pKa = 6.95ISVANGVLTLTATPTSGQGTSSEE88 pKa = 4.52SPHH91 pKa = 5.72LTIDD95 pKa = 3.71YY96 pKa = 10.5FSGTIYY102 pKa = 10.86AKK104 pKa = 10.74DD105 pKa = 4.65DD106 pKa = 3.41ITVDD110 pKa = 3.23GTDD113 pKa = 3.1VVGYY117 pKa = 9.1VVEE120 pKa = 4.71GEE122 pKa = 4.9FIAPTAVGTWPAFWLTAVNSWPPEE146 pKa = 3.67ADD148 pKa = 2.63IGEE151 pKa = 4.35WKK153 pKa = 9.01GTQQTWFNTFNTSSEE168 pKa = 4.27VKK170 pKa = 9.03STIVAWPDD178 pKa = 3.04DD179 pKa = 4.25GEE181 pKa = 4.3FHH183 pKa = 6.73SVKK186 pKa = 10.51AVLSSIEE193 pKa = 4.15GDD195 pKa = 3.63SSDD198 pKa = 3.67VTIDD202 pKa = 3.63YY203 pKa = 11.21YY204 pKa = 11.46LDD206 pKa = 3.42NVLQATHH213 pKa = 6.19VGADD217 pKa = 3.57FLGAPLYY224 pKa = 10.65LIIDD228 pKa = 4.24LQMEE232 pKa = 5.0GSSGSPGPTGTTTYY246 pKa = 10.51QIRR249 pKa = 11.84NVQVTKK255 pKa = 10.12VTSS258 pKa = 3.3
Molecular weight: 27.71 kDa
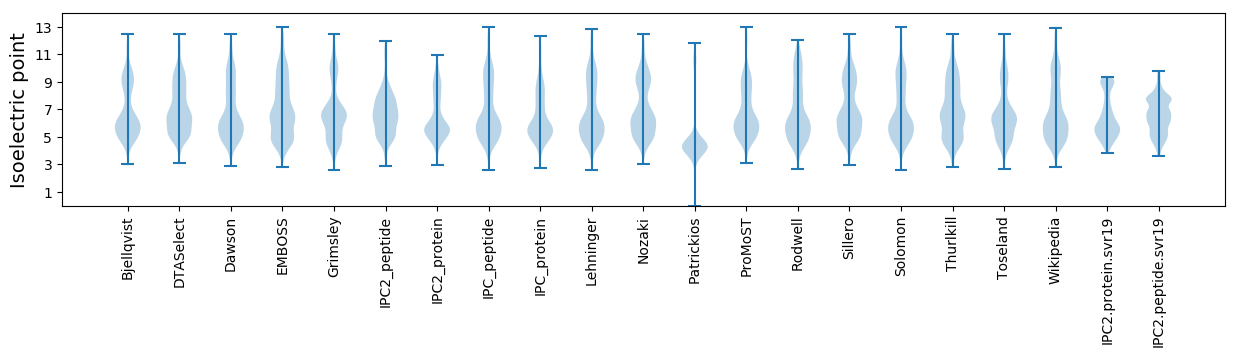
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7S1Z8|R7S1Z8_PUNST Uncharacterized protein OS=Punctularia strigosozonata (strain HHB-11173) OX=741275 GN=PUNSTDRAFT_139108 PE=4 SV=1
MM1 pKa = 8.07RR2 pKa = 11.84SGACCAPLWPVQGGLRR18 pKa = 11.84WTACDD23 pKa = 3.06RR24 pKa = 11.84RR25 pKa = 11.84LRR27 pKa = 11.84EE28 pKa = 4.12LGRR31 pKa = 11.84LKK33 pKa = 10.57PRR35 pKa = 11.84IAIALHH41 pKa = 5.15STWSRR46 pKa = 11.84VYY48 pKa = 10.7GGLGVALSIKK58 pKa = 10.33DD59 pKa = 3.59ALVWTQTSKK68 pKa = 10.85HH69 pKa = 5.58YY70 pKa = 10.49NQSLDD75 pKa = 3.4FRR77 pKa = 11.84THH79 pKa = 7.04CSTTSHH85 pKa = 7.07LAFLPCIHH93 pKa = 6.74LHH95 pKa = 6.06FLPSRR100 pKa = 11.84PISHH104 pKa = 7.39PCVILSPTKK113 pKa = 9.38GTAFYY118 pKa = 10.21TRR120 pKa = 11.84VKK122 pKa = 9.93TSPAPLRR129 pKa = 11.84LRR131 pKa = 11.84LSRR134 pKa = 11.84GPSDD138 pKa = 4.36LRR140 pKa = 11.84IDD142 pKa = 3.92RR143 pKa = 11.84LRR145 pKa = 11.84ASPDD149 pKa = 3.22LGPGRR154 pKa = 11.84SAHH157 pKa = 7.6DD158 pKa = 3.64SDD160 pKa = 5.7LDD162 pKa = 3.95NNN164 pKa = 4.06
MM1 pKa = 8.07RR2 pKa = 11.84SGACCAPLWPVQGGLRR18 pKa = 11.84WTACDD23 pKa = 3.06RR24 pKa = 11.84RR25 pKa = 11.84LRR27 pKa = 11.84EE28 pKa = 4.12LGRR31 pKa = 11.84LKK33 pKa = 10.57PRR35 pKa = 11.84IAIALHH41 pKa = 5.15STWSRR46 pKa = 11.84VYY48 pKa = 10.7GGLGVALSIKK58 pKa = 10.33DD59 pKa = 3.59ALVWTQTSKK68 pKa = 10.85HH69 pKa = 5.58YY70 pKa = 10.49NQSLDD75 pKa = 3.4FRR77 pKa = 11.84THH79 pKa = 7.04CSTTSHH85 pKa = 7.07LAFLPCIHH93 pKa = 6.74LHH95 pKa = 6.06FLPSRR100 pKa = 11.84PISHH104 pKa = 7.39PCVILSPTKK113 pKa = 9.38GTAFYY118 pKa = 10.21TRR120 pKa = 11.84VKK122 pKa = 9.93TSPAPLRR129 pKa = 11.84LRR131 pKa = 11.84LSRR134 pKa = 11.84GPSDD138 pKa = 4.36LRR140 pKa = 11.84IDD142 pKa = 3.92RR143 pKa = 11.84LRR145 pKa = 11.84ASPDD149 pKa = 3.22LGPGRR154 pKa = 11.84SAHH157 pKa = 7.6DD158 pKa = 3.64SDD160 pKa = 5.7LDD162 pKa = 3.95NNN164 pKa = 4.06
Molecular weight: 18.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
867311 |
50 |
3615 |
414.4 |
45.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.641 ± 0.052 | 1.321 ± 0.022 |
5.892 ± 0.043 | 5.788 ± 0.063 |
3.493 ± 0.032 | 6.831 ± 0.056 |
2.634 ± 0.025 | 4.484 ± 0.04 |
4.028 ± 0.051 | 8.98 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.007 ± 0.022 | 3.148 ± 0.033 |
6.686 ± 0.069 | 3.58 ± 0.034 |
6.602 ± 0.054 | 8.217 ± 0.065 |
5.989 ± 0.045 | 6.537 ± 0.04 |
1.544 ± 0.023 | 2.598 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
