
Bromus-associated circular DNA virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
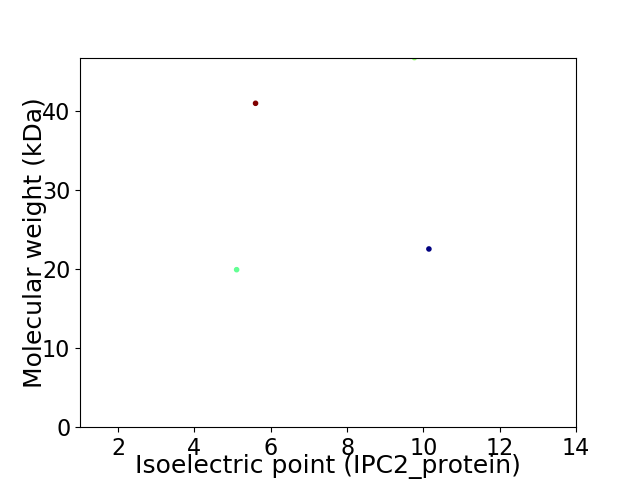
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
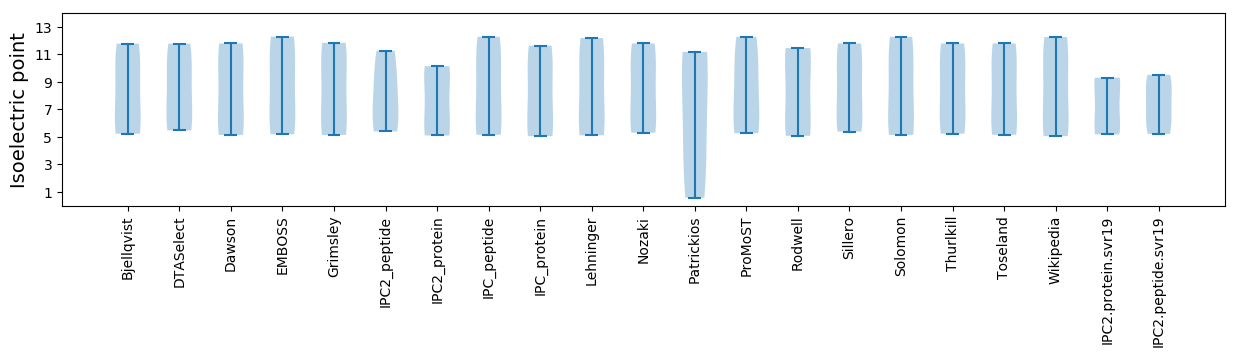
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4U9K0|A0A0B4U9K0_9VIRU Uncharacterized protein OS=Bromus-associated circular DNA virus 1 OX=1590154 PE=4 SV=1
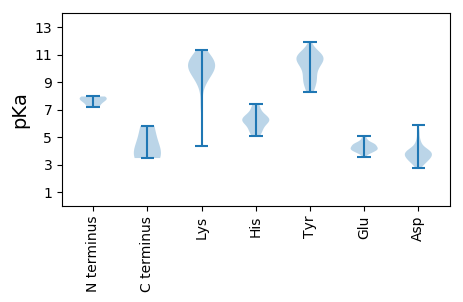
MM1 pKa = 7.82ASGLPGEE8 pKa = 4.48HH9 pKa = 6.62SGDD12 pKa = 3.3GGEE15 pKa = 3.97RR16 pKa = 11.84SVVARR21 pKa = 11.84HH22 pKa = 5.63VPDD25 pKa = 3.69EE26 pKa = 4.23QVCGQSPEE34 pKa = 4.12IRR36 pKa = 11.84SGDD39 pKa = 3.44QKK41 pKa = 11.28LRR43 pKa = 11.84GAGRR47 pKa = 11.84YY48 pKa = 8.85LDD50 pKa = 4.06YY51 pKa = 11.11IRR53 pKa = 11.84LCFAAWCCSEE63 pKa = 4.62HH64 pKa = 6.17PQSGFCEE71 pKa = 4.1PVYY74 pKa = 10.93QPDD77 pKa = 2.8QRR79 pKa = 11.84LAYY82 pKa = 9.63RR83 pKa = 11.84PSEE86 pKa = 4.04RR87 pKa = 11.84VQSGGACGLPEE98 pKa = 4.7SDD100 pKa = 3.47VRR102 pKa = 11.84FAGYY106 pKa = 10.08HH107 pKa = 6.14PLSQYY112 pKa = 11.89ALLQTVQDD120 pKa = 4.01QEE122 pKa = 4.28DD123 pKa = 4.11HH124 pKa = 6.18QGHH127 pKa = 7.4PGTGSGASPPCYY139 pKa = 9.59RR140 pKa = 11.84EE141 pKa = 3.74AAEE144 pKa = 4.45HH145 pKa = 5.8VRR147 pKa = 11.84HH148 pKa = 6.27SDD150 pKa = 3.0VGQCEE155 pKa = 4.04FCAVYY160 pKa = 10.17GGPGLPYY167 pKa = 10.46PGDD170 pKa = 3.16FWVYY174 pKa = 8.97PDD176 pKa = 4.22YY177 pKa = 11.36GAGQHH182 pKa = 5.25WW183 pKa = 3.56
MM1 pKa = 7.82ASGLPGEE8 pKa = 4.48HH9 pKa = 6.62SGDD12 pKa = 3.3GGEE15 pKa = 3.97RR16 pKa = 11.84SVVARR21 pKa = 11.84HH22 pKa = 5.63VPDD25 pKa = 3.69EE26 pKa = 4.23QVCGQSPEE34 pKa = 4.12IRR36 pKa = 11.84SGDD39 pKa = 3.44QKK41 pKa = 11.28LRR43 pKa = 11.84GAGRR47 pKa = 11.84YY48 pKa = 8.85LDD50 pKa = 4.06YY51 pKa = 11.11IRR53 pKa = 11.84LCFAAWCCSEE63 pKa = 4.62HH64 pKa = 6.17PQSGFCEE71 pKa = 4.1PVYY74 pKa = 10.93QPDD77 pKa = 2.8QRR79 pKa = 11.84LAYY82 pKa = 9.63RR83 pKa = 11.84PSEE86 pKa = 4.04RR87 pKa = 11.84VQSGGACGLPEE98 pKa = 4.7SDD100 pKa = 3.47VRR102 pKa = 11.84FAGYY106 pKa = 10.08HH107 pKa = 6.14PLSQYY112 pKa = 11.89ALLQTVQDD120 pKa = 4.01QEE122 pKa = 4.28DD123 pKa = 4.11HH124 pKa = 6.18QGHH127 pKa = 7.4PGTGSGASPPCYY139 pKa = 9.59RR140 pKa = 11.84EE141 pKa = 3.74AAEE144 pKa = 4.45HH145 pKa = 5.8VRR147 pKa = 11.84HH148 pKa = 6.27SDD150 pKa = 3.0VGQCEE155 pKa = 4.04FCAVYY160 pKa = 10.17GGPGLPYY167 pKa = 10.46PGDD170 pKa = 3.16FWVYY174 pKa = 8.97PDD176 pKa = 4.22YY177 pKa = 11.36GAGQHH182 pKa = 5.25WW183 pKa = 3.56
Molecular weight: 19.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4U8V1|A0A0B4U8V1_9VIRU Replication-associated protein OS=Bromus-associated circular DNA virus 1 OX=1590154 PE=3 SV=1
MM1 pKa = 7.65RR2 pKa = 11.84AFAFRR7 pKa = 11.84GAATTLGFIADD18 pKa = 4.15NLRR21 pKa = 11.84GAHH24 pKa = 5.05TAYY27 pKa = 10.84RR28 pKa = 11.84MAKK31 pKa = 7.16GTSYY35 pKa = 10.83GRR37 pKa = 11.84SRR39 pKa = 11.84RR40 pKa = 11.84IPARR44 pKa = 11.84NAKK47 pKa = 9.98SLGIGSRR54 pKa = 11.84SKK56 pKa = 9.96RR57 pKa = 11.84TAAVSKK63 pKa = 9.15FRR65 pKa = 11.84RR66 pKa = 11.84VKK68 pKa = 10.42RR69 pKa = 11.84VVVKK73 pKa = 9.38PKK75 pKa = 10.02RR76 pKa = 11.84KK77 pKa = 9.91SFTRR81 pKa = 11.84TKK83 pKa = 10.68RR84 pKa = 11.84RR85 pKa = 11.84TSYY88 pKa = 10.18KK89 pKa = 9.65AGRR92 pKa = 11.84GGFSSALAKK101 pKa = 10.14TGGSSFSIGSGAKK114 pKa = 10.12LYY116 pKa = 10.84RR117 pKa = 11.84GFKK120 pKa = 8.93WAQQPVTFTSTTTQRR135 pKa = 11.84VTGAAGKK142 pKa = 9.59QAVAVLPTYY151 pKa = 10.49RR152 pKa = 11.84INPGANDD159 pKa = 3.85PTNDD163 pKa = 3.26TALIGTNTLDD173 pKa = 4.07SVLLDD178 pKa = 3.49RR179 pKa = 11.84AEE181 pKa = 4.52RR182 pKa = 11.84WLQAYY187 pKa = 8.88QEE189 pKa = 4.5STPATAASAQSSPGMYY205 pKa = 8.28QTNKK209 pKa = 9.91FVVNHH214 pKa = 6.49LKK216 pKa = 10.36YY217 pKa = 10.81DD218 pKa = 3.53QAIRR222 pKa = 11.84NCEE225 pKa = 4.07VQDD228 pKa = 3.74VTLTIYY234 pKa = 10.95DD235 pKa = 3.71CVLRR239 pKa = 11.84PGVAPNIRR247 pKa = 11.84SPGSVNQSISPINDD261 pKa = 2.74WHH263 pKa = 7.34IGLQNEE269 pKa = 4.51YY270 pKa = 10.91NPAAPVAYY278 pKa = 10.39LSQTFDD284 pKa = 3.57SPATTPFHH292 pKa = 6.09STHH295 pKa = 5.64FCKK298 pKa = 10.47LYY300 pKa = 10.47KK301 pKa = 9.83IKK303 pKa = 9.79KK304 pKa = 4.37TTKK307 pKa = 8.45VTLAPGAVHH316 pKa = 6.25HH317 pKa = 6.25HH318 pKa = 6.35HH319 pKa = 6.14VTVKK323 pKa = 8.88PRR325 pKa = 11.84NMFDD329 pKa = 3.19TQTSGNVNSALYY341 pKa = 9.04TADD344 pKa = 3.57RR345 pKa = 11.84GYY347 pKa = 10.72LIPGISGFTLITVLGSIGDD366 pKa = 3.79SAANHH371 pKa = 6.08LLIGTTKK378 pKa = 9.5TAVDD382 pKa = 3.86VVTTTMASFSQFSRR396 pKa = 11.84EE397 pKa = 3.95RR398 pKa = 11.84RR399 pKa = 11.84MHH401 pKa = 5.62MAFDD405 pKa = 4.55GLTQATDD412 pKa = 3.79LQAVQPDD419 pKa = 3.69TGTIQMDD426 pKa = 3.82TTANEE431 pKa = 3.83AA432 pKa = 3.47
MM1 pKa = 7.65RR2 pKa = 11.84AFAFRR7 pKa = 11.84GAATTLGFIADD18 pKa = 4.15NLRR21 pKa = 11.84GAHH24 pKa = 5.05TAYY27 pKa = 10.84RR28 pKa = 11.84MAKK31 pKa = 7.16GTSYY35 pKa = 10.83GRR37 pKa = 11.84SRR39 pKa = 11.84RR40 pKa = 11.84IPARR44 pKa = 11.84NAKK47 pKa = 9.98SLGIGSRR54 pKa = 11.84SKK56 pKa = 9.96RR57 pKa = 11.84TAAVSKK63 pKa = 9.15FRR65 pKa = 11.84RR66 pKa = 11.84VKK68 pKa = 10.42RR69 pKa = 11.84VVVKK73 pKa = 9.38PKK75 pKa = 10.02RR76 pKa = 11.84KK77 pKa = 9.91SFTRR81 pKa = 11.84TKK83 pKa = 10.68RR84 pKa = 11.84RR85 pKa = 11.84TSYY88 pKa = 10.18KK89 pKa = 9.65AGRR92 pKa = 11.84GGFSSALAKK101 pKa = 10.14TGGSSFSIGSGAKK114 pKa = 10.12LYY116 pKa = 10.84RR117 pKa = 11.84GFKK120 pKa = 8.93WAQQPVTFTSTTTQRR135 pKa = 11.84VTGAAGKK142 pKa = 9.59QAVAVLPTYY151 pKa = 10.49RR152 pKa = 11.84INPGANDD159 pKa = 3.85PTNDD163 pKa = 3.26TALIGTNTLDD173 pKa = 4.07SVLLDD178 pKa = 3.49RR179 pKa = 11.84AEE181 pKa = 4.52RR182 pKa = 11.84WLQAYY187 pKa = 8.88QEE189 pKa = 4.5STPATAASAQSSPGMYY205 pKa = 8.28QTNKK209 pKa = 9.91FVVNHH214 pKa = 6.49LKK216 pKa = 10.36YY217 pKa = 10.81DD218 pKa = 3.53QAIRR222 pKa = 11.84NCEE225 pKa = 4.07VQDD228 pKa = 3.74VTLTIYY234 pKa = 10.95DD235 pKa = 3.71CVLRR239 pKa = 11.84PGVAPNIRR247 pKa = 11.84SPGSVNQSISPINDD261 pKa = 2.74WHH263 pKa = 7.34IGLQNEE269 pKa = 4.51YY270 pKa = 10.91NPAAPVAYY278 pKa = 10.39LSQTFDD284 pKa = 3.57SPATTPFHH292 pKa = 6.09STHH295 pKa = 5.64FCKK298 pKa = 10.47LYY300 pKa = 10.47KK301 pKa = 9.83IKK303 pKa = 9.79KK304 pKa = 4.37TTKK307 pKa = 8.45VTLAPGAVHH316 pKa = 6.25HH317 pKa = 6.25HH318 pKa = 6.35HH319 pKa = 6.14VTVKK323 pKa = 8.88PRR325 pKa = 11.84NMFDD329 pKa = 3.19TQTSGNVNSALYY341 pKa = 9.04TADD344 pKa = 3.57RR345 pKa = 11.84GYY347 pKa = 10.72LIPGISGFTLITVLGSIGDD366 pKa = 3.79SAANHH371 pKa = 6.08LLIGTTKK378 pKa = 9.5TAVDD382 pKa = 3.86VVTTTMASFSQFSRR396 pKa = 11.84EE397 pKa = 3.95RR398 pKa = 11.84RR399 pKa = 11.84MHH401 pKa = 5.62MAFDD405 pKa = 4.55GLTQATDD412 pKa = 3.79LQAVQPDD419 pKa = 3.69TGTIQMDD426 pKa = 3.82TTANEE431 pKa = 3.83AA432 pKa = 3.47
Molecular weight: 46.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1180 |
183 |
432 |
295.0 |
32.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.644 ± 1.149 | 2.119 ± 0.66 |
5.763 ± 0.836 | 3.39 ± 0.914 |
4.237 ± 0.93 | 8.305 ± 1.282 |
2.966 ± 0.4 | 4.322 ± 0.885 |
3.898 ± 0.82 | 7.712 ± 1.167 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.271 ± 0.248 | 3.305 ± 0.676 |
5.339 ± 0.791 | 4.831 ± 0.562 |
8.475 ± 1.646 | 6.949 ± 0.859 |
7.119 ± 2.028 | 7.203 ± 1.507 |
1.017 ± 0.276 | 3.136 ± 0.676 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
