
Hubei tombus-like virus 24
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
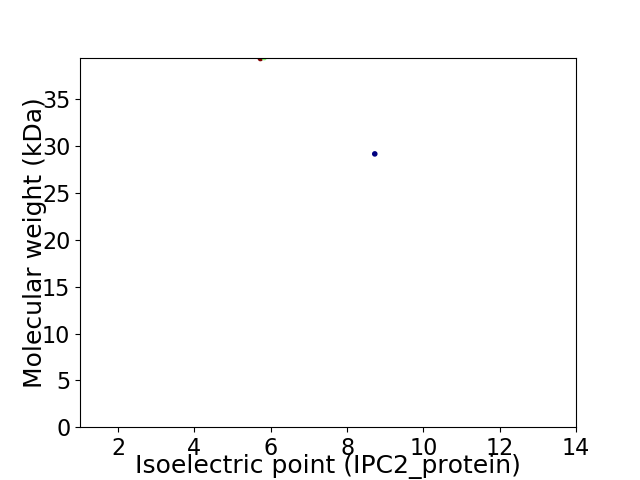
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KG46|A0A1L3KG46_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 24 OX=1923271 PE=4 SV=1
MM1 pKa = 7.55FIKK4 pKa = 10.36HH5 pKa = 5.99EE6 pKa = 4.62KK7 pKa = 10.47GDD9 pKa = 4.12DD10 pKa = 3.74PEE12 pKa = 4.32STPRR16 pKa = 11.84GIQYY20 pKa = 10.63RR21 pKa = 11.84SFKK24 pKa = 8.17WTALFKK30 pKa = 10.57RR31 pKa = 11.84VLGPFEE37 pKa = 3.81MRR39 pKa = 11.84LWRR42 pKa = 11.84LGQDD46 pKa = 3.89FQPSPLAEE54 pKa = 3.82RR55 pKa = 11.84MFSKK59 pKa = 10.51NLNPRR64 pKa = 11.84AVAHH68 pKa = 5.9NLRR71 pKa = 11.84NGWLKK76 pKa = 10.52FADD79 pKa = 4.06PVADD83 pKa = 3.49LWDD86 pKa = 3.77VSRR89 pKa = 11.84MDD91 pKa = 3.15AHH93 pKa = 7.28LGRR96 pKa = 11.84LVRR99 pKa = 11.84EE100 pKa = 4.18MIEE103 pKa = 4.06FPTYY107 pKa = 11.03RR108 pKa = 11.84MGTYY112 pKa = 10.59GFDD115 pKa = 3.68DD116 pKa = 5.24FIGAMRR122 pKa = 11.84HH123 pKa = 5.3NICRR127 pKa = 11.84TKK129 pKa = 10.89NGIVYY134 pKa = 10.15EE135 pKa = 4.28MDD137 pKa = 2.99YY138 pKa = 11.03TMCSGEE144 pKa = 4.23ACTSAGDD151 pKa = 4.39SIVMAAVLDD160 pKa = 4.09FVYY163 pKa = 10.75RR164 pKa = 11.84DD165 pKa = 3.58IPHH168 pKa = 6.8HH169 pKa = 6.54KK170 pKa = 9.67LVCGDD175 pKa = 3.69DD176 pKa = 3.47CVVIRR181 pKa = 11.84EE182 pKa = 4.27RR183 pKa = 11.84GCVPDD188 pKa = 3.51SSVFAQCGLPVKK200 pKa = 10.52SDD202 pKa = 3.28VVDD205 pKa = 3.19QFEE208 pKa = 4.18RR209 pKa = 11.84VEE211 pKa = 4.55FCQSRR216 pKa = 11.84PVCVAGVWTMVRR228 pKa = 11.84NPDD231 pKa = 3.3RR232 pKa = 11.84VVNRR236 pKa = 11.84ALCTIKK242 pKa = 10.88NFGGEE247 pKa = 3.95VRR249 pKa = 11.84PYY251 pKa = 10.28QDD253 pKa = 2.37WLASVGVGEE262 pKa = 4.81LEE264 pKa = 4.27SGAGVPVVQTLALEE278 pKa = 4.12LMKK281 pKa = 10.69LGQPRR286 pKa = 11.84PHH288 pKa = 6.9FVRR291 pKa = 11.84EE292 pKa = 4.06YY293 pKa = 9.94LEE295 pKa = 3.88HH296 pKa = 7.38RR297 pKa = 11.84RR298 pKa = 11.84TLQQRR303 pKa = 11.84EE304 pKa = 4.2PLEE307 pKa = 4.21VTDD310 pKa = 4.05EE311 pKa = 4.32TRR313 pKa = 11.84HH314 pKa = 5.58SFWLAWGWTPEE325 pKa = 3.84EE326 pKa = 3.87QVRR329 pKa = 11.84FEE331 pKa = 4.11SLLLGGAEE339 pKa = 3.97FDD341 pKa = 4.36LVVV344 pKa = 3.48
MM1 pKa = 7.55FIKK4 pKa = 10.36HH5 pKa = 5.99EE6 pKa = 4.62KK7 pKa = 10.47GDD9 pKa = 4.12DD10 pKa = 3.74PEE12 pKa = 4.32STPRR16 pKa = 11.84GIQYY20 pKa = 10.63RR21 pKa = 11.84SFKK24 pKa = 8.17WTALFKK30 pKa = 10.57RR31 pKa = 11.84VLGPFEE37 pKa = 3.81MRR39 pKa = 11.84LWRR42 pKa = 11.84LGQDD46 pKa = 3.89FQPSPLAEE54 pKa = 3.82RR55 pKa = 11.84MFSKK59 pKa = 10.51NLNPRR64 pKa = 11.84AVAHH68 pKa = 5.9NLRR71 pKa = 11.84NGWLKK76 pKa = 10.52FADD79 pKa = 4.06PVADD83 pKa = 3.49LWDD86 pKa = 3.77VSRR89 pKa = 11.84MDD91 pKa = 3.15AHH93 pKa = 7.28LGRR96 pKa = 11.84LVRR99 pKa = 11.84EE100 pKa = 4.18MIEE103 pKa = 4.06FPTYY107 pKa = 11.03RR108 pKa = 11.84MGTYY112 pKa = 10.59GFDD115 pKa = 3.68DD116 pKa = 5.24FIGAMRR122 pKa = 11.84HH123 pKa = 5.3NICRR127 pKa = 11.84TKK129 pKa = 10.89NGIVYY134 pKa = 10.15EE135 pKa = 4.28MDD137 pKa = 2.99YY138 pKa = 11.03TMCSGEE144 pKa = 4.23ACTSAGDD151 pKa = 4.39SIVMAAVLDD160 pKa = 4.09FVYY163 pKa = 10.75RR164 pKa = 11.84DD165 pKa = 3.58IPHH168 pKa = 6.8HH169 pKa = 6.54KK170 pKa = 9.67LVCGDD175 pKa = 3.69DD176 pKa = 3.47CVVIRR181 pKa = 11.84EE182 pKa = 4.27RR183 pKa = 11.84GCVPDD188 pKa = 3.51SSVFAQCGLPVKK200 pKa = 10.52SDD202 pKa = 3.28VVDD205 pKa = 3.19QFEE208 pKa = 4.18RR209 pKa = 11.84VEE211 pKa = 4.55FCQSRR216 pKa = 11.84PVCVAGVWTMVRR228 pKa = 11.84NPDD231 pKa = 3.3RR232 pKa = 11.84VVNRR236 pKa = 11.84ALCTIKK242 pKa = 10.88NFGGEE247 pKa = 3.95VRR249 pKa = 11.84PYY251 pKa = 10.28QDD253 pKa = 2.37WLASVGVGEE262 pKa = 4.81LEE264 pKa = 4.27SGAGVPVVQTLALEE278 pKa = 4.12LMKK281 pKa = 10.69LGQPRR286 pKa = 11.84PHH288 pKa = 6.9FVRR291 pKa = 11.84EE292 pKa = 4.06YY293 pKa = 9.94LEE295 pKa = 3.88HH296 pKa = 7.38RR297 pKa = 11.84RR298 pKa = 11.84TLQQRR303 pKa = 11.84EE304 pKa = 4.2PLEE307 pKa = 4.21VTDD310 pKa = 4.05EE311 pKa = 4.32TRR313 pKa = 11.84HH314 pKa = 5.58SFWLAWGWTPEE325 pKa = 3.84EE326 pKa = 3.87QVRR329 pKa = 11.84FEE331 pKa = 4.11SLLLGGAEE339 pKa = 3.97FDD341 pKa = 4.36LVVV344 pKa = 3.48
Molecular weight: 39.37 kDa
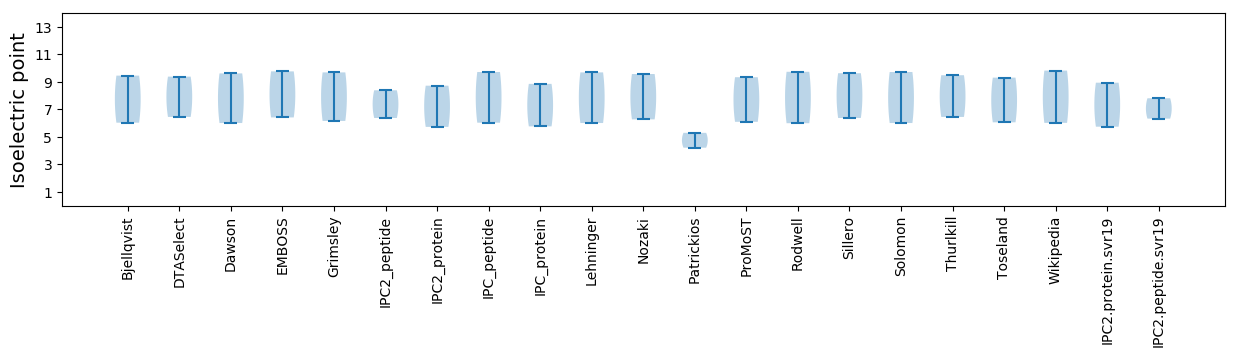
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG46|A0A1L3KG46_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 24 OX=1923271 PE=4 SV=1
MM1 pKa = 7.84GIGLSDD7 pKa = 3.85SSRR10 pKa = 11.84STPSLCATDD19 pKa = 5.29AGVCHH24 pKa = 6.34QSPQNPVARR33 pKa = 11.84ISGGRR38 pKa = 11.84DD39 pKa = 2.62EE40 pKa = 4.86RR41 pKa = 11.84RR42 pKa = 11.84PVNRR46 pKa = 11.84RR47 pKa = 11.84GVPVATQTSEE57 pKa = 4.27LPGHH61 pKa = 6.36ARR63 pKa = 11.84SVDD66 pKa = 3.41EE67 pKa = 4.13SHH69 pKa = 7.58PARR72 pKa = 11.84DD73 pKa = 3.73VVSKK77 pKa = 10.74SVAAIQDD84 pKa = 3.61RR85 pKa = 11.84VDD87 pKa = 3.31GLGAEE92 pKa = 4.56LSLPGWQRR100 pKa = 11.84AIPDD104 pKa = 3.94PRR106 pKa = 11.84KK107 pKa = 9.17DD108 pKa = 3.32TYY110 pKa = 11.57SEE112 pKa = 3.98LFSYY116 pKa = 10.61LRR118 pKa = 11.84RR119 pKa = 11.84QCGTSRR125 pKa = 11.84ISMLTLDD132 pKa = 3.89SMRR135 pKa = 11.84RR136 pKa = 11.84MGDD139 pKa = 2.24SWLARR144 pKa = 11.84NRR146 pKa = 11.84VWLSEE151 pKa = 4.03TARR154 pKa = 11.84TDD156 pKa = 3.06VLEE159 pKa = 4.02RR160 pKa = 11.84VIVDD164 pKa = 3.81LAQPQKK170 pKa = 11.09SDD172 pKa = 3.14TYY174 pKa = 10.65LASHH178 pKa = 5.3VTKK181 pKa = 9.73GTFYY185 pKa = 11.2SPISLGVGDD194 pKa = 3.86IKK196 pKa = 11.3SRR198 pKa = 11.84IAHH201 pKa = 5.84ANEE204 pKa = 3.28LRR206 pKa = 11.84AGRR209 pKa = 11.84RR210 pKa = 11.84WVPTKK215 pKa = 10.84NSWHH219 pKa = 6.46HH220 pKa = 5.81VLAALLFLAVALAHH234 pKa = 6.63PIGLVIALVGLGGVGYY250 pKa = 10.1SWVSTDD256 pKa = 2.81HH257 pKa = 6.73GLWEE261 pKa = 3.92QVIPEE266 pKa = 4.21KK267 pKa = 11.0
MM1 pKa = 7.84GIGLSDD7 pKa = 3.85SSRR10 pKa = 11.84STPSLCATDD19 pKa = 5.29AGVCHH24 pKa = 6.34QSPQNPVARR33 pKa = 11.84ISGGRR38 pKa = 11.84DD39 pKa = 2.62EE40 pKa = 4.86RR41 pKa = 11.84RR42 pKa = 11.84PVNRR46 pKa = 11.84RR47 pKa = 11.84GVPVATQTSEE57 pKa = 4.27LPGHH61 pKa = 6.36ARR63 pKa = 11.84SVDD66 pKa = 3.41EE67 pKa = 4.13SHH69 pKa = 7.58PARR72 pKa = 11.84DD73 pKa = 3.73VVSKK77 pKa = 10.74SVAAIQDD84 pKa = 3.61RR85 pKa = 11.84VDD87 pKa = 3.31GLGAEE92 pKa = 4.56LSLPGWQRR100 pKa = 11.84AIPDD104 pKa = 3.94PRR106 pKa = 11.84KK107 pKa = 9.17DD108 pKa = 3.32TYY110 pKa = 11.57SEE112 pKa = 3.98LFSYY116 pKa = 10.61LRR118 pKa = 11.84RR119 pKa = 11.84QCGTSRR125 pKa = 11.84ISMLTLDD132 pKa = 3.89SMRR135 pKa = 11.84RR136 pKa = 11.84MGDD139 pKa = 2.24SWLARR144 pKa = 11.84NRR146 pKa = 11.84VWLSEE151 pKa = 4.03TARR154 pKa = 11.84TDD156 pKa = 3.06VLEE159 pKa = 4.02RR160 pKa = 11.84VIVDD164 pKa = 3.81LAQPQKK170 pKa = 11.09SDD172 pKa = 3.14TYY174 pKa = 10.65LASHH178 pKa = 5.3VTKK181 pKa = 9.73GTFYY185 pKa = 11.2SPISLGVGDD194 pKa = 3.86IKK196 pKa = 11.3SRR198 pKa = 11.84IAHH201 pKa = 5.84ANEE204 pKa = 3.28LRR206 pKa = 11.84AGRR209 pKa = 11.84RR210 pKa = 11.84WVPTKK215 pKa = 10.84NSWHH219 pKa = 6.46HH220 pKa = 5.81VLAALLFLAVALAHH234 pKa = 6.63PIGLVIALVGLGGVGYY250 pKa = 10.1SWVSTDD256 pKa = 2.81HH257 pKa = 6.73GLWEE261 pKa = 3.92QVIPEE266 pKa = 4.21KK267 pKa = 11.0
Molecular weight: 29.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
611 |
267 |
344 |
305.5 |
34.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.038 ± 1.063 | 2.128 ± 0.677 |
6.219 ± 0.153 | 5.565 ± 1.227 |
3.601 ± 1.67 | 7.856 ± 0.259 |
2.946 ± 0.286 | 3.601 ± 0.603 |
2.946 ± 0.219 | 9.002 ± 0.496 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.619 ± 0.756 | 2.291 ± 0.282 |
5.565 ± 0.036 | 3.437 ± 0.045 |
9.002 ± 0.244 | 7.201 ± 2.215 |
4.583 ± 0.446 | 9.656 ± 0.45 |
2.619 ± 0.002 | 2.128 ± 0.172 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
