
Sulfolobales Mexican rudivirus 1
Taxonomy: Viruses; Adnaviria; Zilligvirae; Taleaviricota; Tokiviricetes; Ligamenvirales; Rudiviridae; Rudivirus; unclassified Rudivirus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
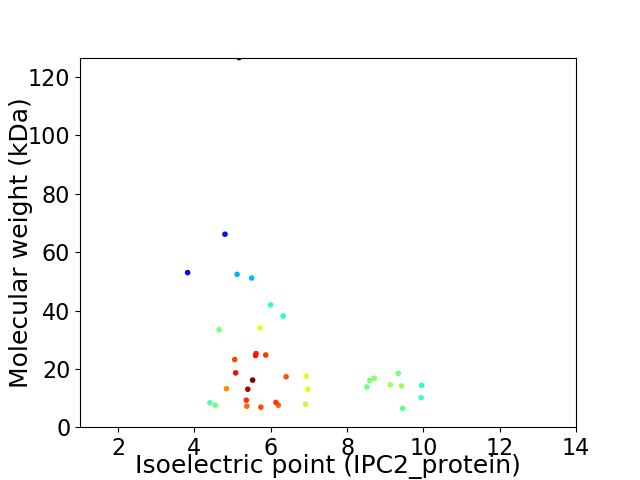
Virtual 2D-PAGE plot for 37 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
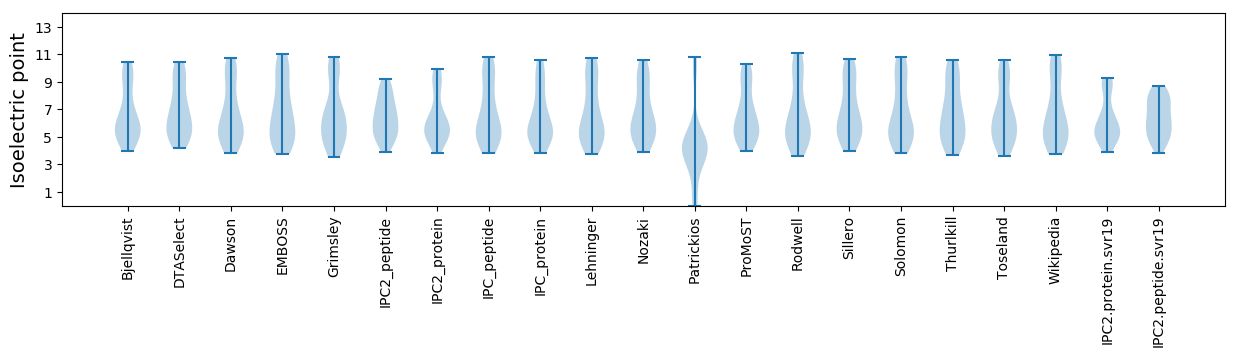
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4NZG9|K4NZG9_9VIRU Uncharacterized protein OS=Sulfolobales Mexican rudivirus 1 OX=1245909 PE=4 SV=1
MM1 pKa = 6.91VTLQQVSSVTVTSPWSDD18 pKa = 3.52YY19 pKa = 11.71NEE21 pKa = 4.24MPNQIQYY28 pKa = 11.16GSIVNPFYY36 pKa = 10.56PVFIFSYY43 pKa = 11.08VNTSNCGLWVVGYY56 pKa = 10.37VSGDD60 pKa = 3.17NAGVFFNHH68 pKa = 6.08ACGWVNVSGMFYY80 pKa = 10.52FDD82 pKa = 3.56AFGYY86 pKa = 8.81PYY88 pKa = 11.45AMGGGYY94 pKa = 8.98MSGVSSCANNGAVWWTFVNQKK115 pKa = 8.67TGSVNAYY122 pKa = 7.3CTGLIPNNDD131 pKa = 2.78VSMDD135 pKa = 4.5FISAVADD142 pKa = 3.9LNQQNILLLSLDD154 pKa = 3.41ISVMAFLIWIIPVSEE169 pKa = 4.35ISTLLNNSSPPSIKK183 pKa = 10.09FAVVKK188 pKa = 8.33TTDD191 pKa = 2.97VPNQYY196 pKa = 8.31MQQFFPVIYY205 pKa = 10.02DD206 pKa = 3.34GNLIIGSTYY215 pKa = 10.85DD216 pKa = 3.36YY217 pKa = 11.03VAYY220 pKa = 10.07LAVMPLSEE228 pKa = 4.3IYY230 pKa = 10.32VNAMTGIPTTNTPATYY246 pKa = 10.64VGTQYY251 pKa = 10.77TYY253 pKa = 10.99SEE255 pKa = 4.82YY256 pKa = 11.02EE257 pKa = 4.01FVTWPIITTIAIPSSGGLSIYY278 pKa = 10.22VSLMLGAKK286 pKa = 10.02NYY288 pKa = 10.64LLYY291 pKa = 10.24IANLSPSNWSVVSSFTFYY309 pKa = 10.41LTDD312 pKa = 3.37VASSICTAPVGSTWNGVFILMFNGPSSSCNTLYY345 pKa = 11.06VVVADD350 pKa = 4.67PKK352 pKa = 10.13SSNYY356 pKa = 9.88EE357 pKa = 3.5YY358 pKa = 10.34TYY360 pKa = 11.3INMTQNTDD368 pKa = 2.96IAILGYY374 pKa = 10.09EE375 pKa = 4.62GYY377 pKa = 10.55LVVSQQPLSDD387 pKa = 3.58STVTFVVYY395 pKa = 10.29QILLDD400 pKa = 3.57HH401 pKa = 6.78TYY403 pKa = 10.38VFQNVAVSTTSSTITFSGVLYY424 pKa = 10.54DD425 pKa = 4.06EE426 pKa = 4.66TAGAPVANATVWLVAVRR443 pKa = 11.84SYY445 pKa = 11.14VKK447 pKa = 10.29QYY449 pKa = 11.38SNDD452 pKa = 3.56VVPLASTTTDD462 pKa = 3.03SAGHH466 pKa = 5.15FTVSAPMQSGYY477 pKa = 10.25NAYY480 pKa = 9.96GVLYY484 pKa = 9.78TPP486 pKa = 5.2
MM1 pKa = 6.91VTLQQVSSVTVTSPWSDD18 pKa = 3.52YY19 pKa = 11.71NEE21 pKa = 4.24MPNQIQYY28 pKa = 11.16GSIVNPFYY36 pKa = 10.56PVFIFSYY43 pKa = 11.08VNTSNCGLWVVGYY56 pKa = 10.37VSGDD60 pKa = 3.17NAGVFFNHH68 pKa = 6.08ACGWVNVSGMFYY80 pKa = 10.52FDD82 pKa = 3.56AFGYY86 pKa = 8.81PYY88 pKa = 11.45AMGGGYY94 pKa = 8.98MSGVSSCANNGAVWWTFVNQKK115 pKa = 8.67TGSVNAYY122 pKa = 7.3CTGLIPNNDD131 pKa = 2.78VSMDD135 pKa = 4.5FISAVADD142 pKa = 3.9LNQQNILLLSLDD154 pKa = 3.41ISVMAFLIWIIPVSEE169 pKa = 4.35ISTLLNNSSPPSIKK183 pKa = 10.09FAVVKK188 pKa = 8.33TTDD191 pKa = 2.97VPNQYY196 pKa = 8.31MQQFFPVIYY205 pKa = 10.02DD206 pKa = 3.34GNLIIGSTYY215 pKa = 10.85DD216 pKa = 3.36YY217 pKa = 11.03VAYY220 pKa = 10.07LAVMPLSEE228 pKa = 4.3IYY230 pKa = 10.32VNAMTGIPTTNTPATYY246 pKa = 10.64VGTQYY251 pKa = 10.77TYY253 pKa = 10.99SEE255 pKa = 4.82YY256 pKa = 11.02EE257 pKa = 4.01FVTWPIITTIAIPSSGGLSIYY278 pKa = 10.22VSLMLGAKK286 pKa = 10.02NYY288 pKa = 10.64LLYY291 pKa = 10.24IANLSPSNWSVVSSFTFYY309 pKa = 10.41LTDD312 pKa = 3.37VASSICTAPVGSTWNGVFILMFNGPSSSCNTLYY345 pKa = 11.06VVVADD350 pKa = 4.67PKK352 pKa = 10.13SSNYY356 pKa = 9.88EE357 pKa = 3.5YY358 pKa = 10.34TYY360 pKa = 11.3INMTQNTDD368 pKa = 2.96IAILGYY374 pKa = 10.09EE375 pKa = 4.62GYY377 pKa = 10.55LVVSQQPLSDD387 pKa = 3.58STVTFVVYY395 pKa = 10.29QILLDD400 pKa = 3.57HH401 pKa = 6.78TYY403 pKa = 10.38VFQNVAVSTTSSTITFSGVLYY424 pKa = 10.54DD425 pKa = 4.06EE426 pKa = 4.66TAGAPVANATVWLVAVRR443 pKa = 11.84SYY445 pKa = 11.14VKK447 pKa = 10.29QYY449 pKa = 11.38SNDD452 pKa = 3.56VVPLASTTTDD462 pKa = 3.03SAGHH466 pKa = 5.15FTVSAPMQSGYY477 pKa = 10.25NAYY480 pKa = 9.96GVLYY484 pKa = 9.78TPP486 pKa = 5.2
Molecular weight: 52.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4NX87|K4NX87_9VIRU Putative SAM-dependent methyltransferase OS=Sulfolobales Mexican rudivirus 1 OX=1245909 PE=4 SV=1
MM1 pKa = 7.55HH2 pKa = 7.32NGAQKK7 pKa = 10.48RR8 pKa = 11.84VIKK11 pKa = 9.94LINVIRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.09KK21 pKa = 9.85PPAVRR26 pKa = 11.84TWIFHH31 pKa = 5.57YY32 pKa = 9.58IWHH35 pKa = 6.15GRR37 pKa = 11.84SYY39 pKa = 10.59TIATIAVRR47 pKa = 11.84LTGDD51 pKa = 3.32GALHH55 pKa = 5.25VRR57 pKa = 11.84KK58 pKa = 10.14KK59 pKa = 10.4IEE61 pKa = 3.9ARR63 pKa = 11.84LKK65 pKa = 10.32ALEE68 pKa = 4.35GVNVTTFLDD77 pKa = 3.75GEE79 pKa = 4.57YY80 pKa = 10.2IKK82 pKa = 10.47YY83 pKa = 8.51TLEE86 pKa = 3.86AA87 pKa = 4.73
MM1 pKa = 7.55HH2 pKa = 7.32NGAQKK7 pKa = 10.48RR8 pKa = 11.84VIKK11 pKa = 9.94LINVIRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.09KK21 pKa = 9.85PPAVRR26 pKa = 11.84TWIFHH31 pKa = 5.57YY32 pKa = 9.58IWHH35 pKa = 6.15GRR37 pKa = 11.84SYY39 pKa = 10.59TIATIAVRR47 pKa = 11.84LTGDD51 pKa = 3.32GALHH55 pKa = 5.25VRR57 pKa = 11.84KK58 pKa = 10.14KK59 pKa = 10.4IEE61 pKa = 3.9ARR63 pKa = 11.84LKK65 pKa = 10.32ALEE68 pKa = 4.35GVNVTTFLDD77 pKa = 3.75GEE79 pKa = 4.57YY80 pKa = 10.2IKK82 pKa = 10.47YY83 pKa = 8.51TLEE86 pKa = 3.86AA87 pKa = 4.73
Molecular weight: 10.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7852 |
54 |
1106 |
212.2 |
24.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.221 ± 0.415 | 0.777 ± 0.177 |
4.648 ± 0.366 | 6.011 ± 0.711 |
4.661 ± 0.335 | 5.018 ± 0.347 |
1.617 ± 0.23 | 6.572 ± 0.36 |
5.183 ± 0.477 | 10.15 ± 0.452 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.394 ± 0.177 | 5.323 ± 0.364 |
4.075 ± 0.346 | 4.483 ± 0.416 |
4.457 ± 0.498 | 6.597 ± 0.626 |
5.693 ± 0.347 | 8.482 ± 0.479 |
1.032 ± 0.186 | 5.604 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
