
Delphinus delphis polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Zetapolyomavirus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
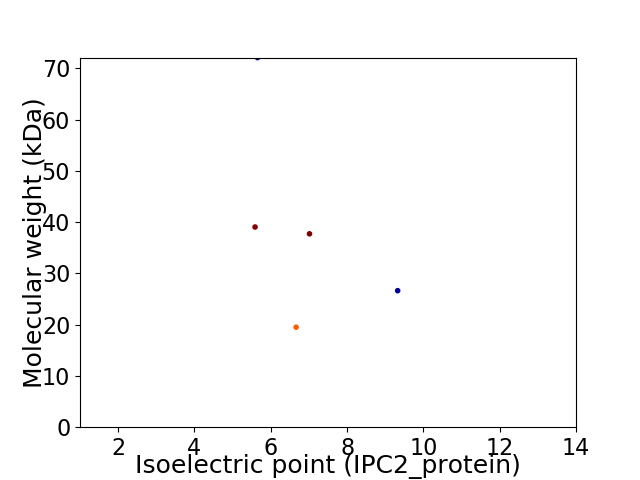
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5M639|S5M639_9POLY Minor capsid protein OS=Delphinus delphis polyomavirus 1 OX=1891756 PE=3 SV=1
MM1 pKa = 7.52SARR4 pKa = 11.84KK5 pKa = 9.68GRR7 pKa = 11.84GAVRR11 pKa = 11.84PPSQVPKK18 pKa = 10.4LIVKK22 pKa = 10.38GGVEE26 pKa = 4.02VLGVKK31 pKa = 9.28TGPDD35 pKa = 3.15SILYY39 pKa = 8.33VEE41 pKa = 5.91CYY43 pKa = 9.4LQPRR47 pKa = 11.84MGDD50 pKa = 3.33PATNGASSGQLTRR63 pKa = 11.84NSTNEE68 pKa = 3.95TDD70 pKa = 4.85NEE72 pKa = 4.47TQQGTRR78 pKa = 11.84LARR81 pKa = 11.84YY82 pKa = 8.92SCGSIQLPLLNDD94 pKa = 3.91DD95 pKa = 4.06MTQGTILMWEE105 pKa = 3.94AFEE108 pKa = 4.92VKK110 pKa = 9.94TEE112 pKa = 3.92LVGINVLTNFHH123 pKa = 6.31SAEE126 pKa = 3.99DD127 pKa = 3.99LAWPDD132 pKa = 3.89GPGLPIQGLNFHH144 pKa = 6.69MFAVGGQPLEE154 pKa = 4.07LQGILMNHH162 pKa = 6.5LTTYY166 pKa = 10.06PDD168 pKa = 3.6DD169 pKa = 3.66VVVPNRR175 pKa = 11.84TNKK178 pKa = 9.08TPSLKK183 pKa = 10.85VLDD186 pKa = 4.24PTAKK190 pKa = 10.34SQLTEE195 pKa = 4.45DD196 pKa = 3.39GKK198 pKa = 11.4FPIEE202 pKa = 4.36CWAPDD207 pKa = 3.61PSKK210 pKa = 11.5NEE212 pKa = 3.61NTRR215 pKa = 11.84YY216 pKa = 9.51FCSLTGGTQTPPVLQQTNTVTTVLLDD242 pKa = 3.84ANGVGPLCKK251 pKa = 9.67GDD253 pKa = 3.66RR254 pKa = 11.84LFVTSADD261 pKa = 3.16IIGIHH266 pKa = 6.84KK267 pKa = 10.04DD268 pKa = 3.34ASHH271 pKa = 6.46HH272 pKa = 5.75AFLRR276 pKa = 11.84GLPRR280 pKa = 11.84FFRR283 pKa = 11.84VGLRR287 pKa = 11.84KK288 pKa = 8.79RR289 pKa = 11.84TVKK292 pKa = 10.55NPYY295 pKa = 9.03PVYY298 pKa = 11.4SMLNSLFTNLNPSITGQSMQGANSQVEE325 pKa = 4.54EE326 pKa = 4.11VSVYY330 pKa = 10.56QGTEE334 pKa = 3.96PLPADD339 pKa = 3.75PDD341 pKa = 3.32MTRR344 pKa = 11.84FINEE348 pKa = 4.53FGQEE352 pKa = 4.21TTSLPHH358 pKa = 6.28
MM1 pKa = 7.52SARR4 pKa = 11.84KK5 pKa = 9.68GRR7 pKa = 11.84GAVRR11 pKa = 11.84PPSQVPKK18 pKa = 10.4LIVKK22 pKa = 10.38GGVEE26 pKa = 4.02VLGVKK31 pKa = 9.28TGPDD35 pKa = 3.15SILYY39 pKa = 8.33VEE41 pKa = 5.91CYY43 pKa = 9.4LQPRR47 pKa = 11.84MGDD50 pKa = 3.33PATNGASSGQLTRR63 pKa = 11.84NSTNEE68 pKa = 3.95TDD70 pKa = 4.85NEE72 pKa = 4.47TQQGTRR78 pKa = 11.84LARR81 pKa = 11.84YY82 pKa = 8.92SCGSIQLPLLNDD94 pKa = 3.91DD95 pKa = 4.06MTQGTILMWEE105 pKa = 3.94AFEE108 pKa = 4.92VKK110 pKa = 9.94TEE112 pKa = 3.92LVGINVLTNFHH123 pKa = 6.31SAEE126 pKa = 3.99DD127 pKa = 3.99LAWPDD132 pKa = 3.89GPGLPIQGLNFHH144 pKa = 6.69MFAVGGQPLEE154 pKa = 4.07LQGILMNHH162 pKa = 6.5LTTYY166 pKa = 10.06PDD168 pKa = 3.6DD169 pKa = 3.66VVVPNRR175 pKa = 11.84TNKK178 pKa = 9.08TPSLKK183 pKa = 10.85VLDD186 pKa = 4.24PTAKK190 pKa = 10.34SQLTEE195 pKa = 4.45DD196 pKa = 3.39GKK198 pKa = 11.4FPIEE202 pKa = 4.36CWAPDD207 pKa = 3.61PSKK210 pKa = 11.5NEE212 pKa = 3.61NTRR215 pKa = 11.84YY216 pKa = 9.51FCSLTGGTQTPPVLQQTNTVTTVLLDD242 pKa = 3.84ANGVGPLCKK251 pKa = 9.67GDD253 pKa = 3.66RR254 pKa = 11.84LFVTSADD261 pKa = 3.16IIGIHH266 pKa = 6.84KK267 pKa = 10.04DD268 pKa = 3.34ASHH271 pKa = 6.46HH272 pKa = 5.75AFLRR276 pKa = 11.84GLPRR280 pKa = 11.84FFRR283 pKa = 11.84VGLRR287 pKa = 11.84KK288 pKa = 8.79RR289 pKa = 11.84TVKK292 pKa = 10.55NPYY295 pKa = 9.03PVYY298 pKa = 11.4SMLNSLFTNLNPSITGQSMQGANSQVEE325 pKa = 4.54EE326 pKa = 4.11VSVYY330 pKa = 10.56QGTEE334 pKa = 3.96PLPADD339 pKa = 3.75PDD341 pKa = 3.32MTRR344 pKa = 11.84FINEE348 pKa = 4.53FGQEE352 pKa = 4.21TTSLPHH358 pKa = 6.28
Molecular weight: 39.07 kDa
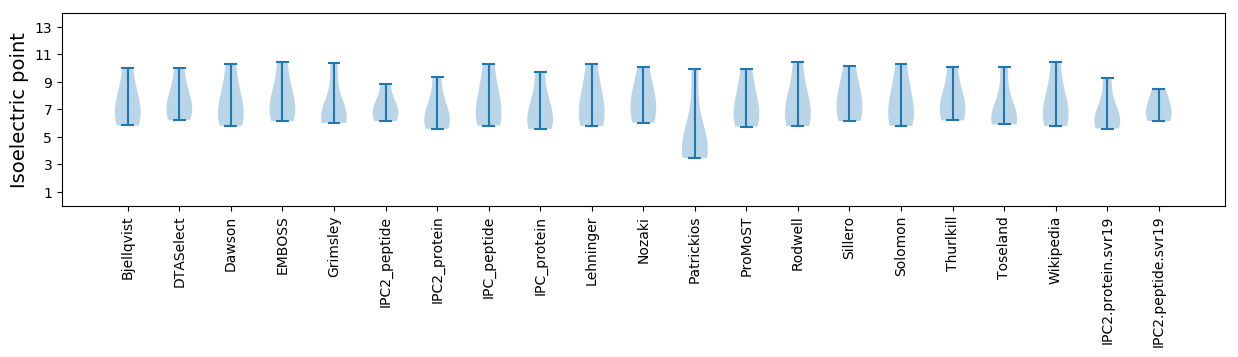
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5MND0|S5MND0_9POLY Small T antigen OS=Delphinus delphis polyomavirus 1 OX=1891756 PE=4 SV=1
MM1 pKa = 7.49ALVEE5 pKa = 4.45WRR7 pKa = 11.84PEE9 pKa = 4.2LIDD12 pKa = 3.52YY13 pKa = 7.92NFPGVRR19 pKa = 11.84WLARR23 pKa = 11.84NVEE26 pKa = 4.28YY27 pKa = 10.54FDD29 pKa = 3.6PRR31 pKa = 11.84FWASEE36 pKa = 3.62LWRR39 pKa = 11.84VFMEE43 pKa = 4.34SVARR47 pKa = 11.84EE48 pKa = 3.7GAAQIEE54 pKa = 4.43AASTALAEE62 pKa = 4.35QGRR65 pKa = 11.84SVAADD70 pKa = 4.5AIARR74 pKa = 11.84SLEE77 pKa = 3.75NANWVISEE85 pKa = 4.3TGALARR91 pKa = 11.84RR92 pKa = 11.84GLDD95 pKa = 3.04VTGALVRR102 pKa = 11.84QAGQRR107 pKa = 11.84AVQSTGSIITNAYY120 pKa = 7.79TNLGDD125 pKa = 3.97YY126 pKa = 10.82YY127 pKa = 10.97RR128 pKa = 11.84GLTPLRR134 pKa = 11.84PPQRR138 pKa = 11.84RR139 pKa = 11.84AMLRR143 pKa = 11.84EE144 pKa = 3.99LEE146 pKa = 4.08KK147 pKa = 10.84SEE149 pKa = 3.96RR150 pKa = 11.84QQTITGEE157 pKa = 4.4VVYY160 pKa = 10.21KK161 pKa = 10.92APAPGGANQRR171 pKa = 11.84ITPEE175 pKa = 3.49WMLPLILGLYY185 pKa = 7.75NHH187 pKa = 6.75PTWGVPDD194 pKa = 3.96AVTALKK200 pKa = 10.59SSQEE204 pKa = 3.89KK205 pKa = 10.31EE206 pKa = 3.98EE207 pKa = 4.1NVSKK211 pKa = 10.57KK212 pKa = 9.96RR213 pKa = 11.84KK214 pKa = 9.11RR215 pKa = 11.84SCEE218 pKa = 3.93TTQPGAKK225 pKa = 8.73TNCKK229 pKa = 9.24RR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84STRR236 pKa = 11.84RR237 pKa = 3.09
MM1 pKa = 7.49ALVEE5 pKa = 4.45WRR7 pKa = 11.84PEE9 pKa = 4.2LIDD12 pKa = 3.52YY13 pKa = 7.92NFPGVRR19 pKa = 11.84WLARR23 pKa = 11.84NVEE26 pKa = 4.28YY27 pKa = 10.54FDD29 pKa = 3.6PRR31 pKa = 11.84FWASEE36 pKa = 3.62LWRR39 pKa = 11.84VFMEE43 pKa = 4.34SVARR47 pKa = 11.84EE48 pKa = 3.7GAAQIEE54 pKa = 4.43AASTALAEE62 pKa = 4.35QGRR65 pKa = 11.84SVAADD70 pKa = 4.5AIARR74 pKa = 11.84SLEE77 pKa = 3.75NANWVISEE85 pKa = 4.3TGALARR91 pKa = 11.84RR92 pKa = 11.84GLDD95 pKa = 3.04VTGALVRR102 pKa = 11.84QAGQRR107 pKa = 11.84AVQSTGSIITNAYY120 pKa = 7.79TNLGDD125 pKa = 3.97YY126 pKa = 10.82YY127 pKa = 10.97RR128 pKa = 11.84GLTPLRR134 pKa = 11.84PPQRR138 pKa = 11.84RR139 pKa = 11.84AMLRR143 pKa = 11.84EE144 pKa = 3.99LEE146 pKa = 4.08KK147 pKa = 10.84SEE149 pKa = 3.96RR150 pKa = 11.84QQTITGEE157 pKa = 4.4VVYY160 pKa = 10.21KK161 pKa = 10.92APAPGGANQRR171 pKa = 11.84ITPEE175 pKa = 3.49WMLPLILGLYY185 pKa = 7.75NHH187 pKa = 6.75PTWGVPDD194 pKa = 3.96AVTALKK200 pKa = 10.59SSQEE204 pKa = 3.89KK205 pKa = 10.31EE206 pKa = 3.98EE207 pKa = 4.1NVSKK211 pKa = 10.57KK212 pKa = 9.96RR213 pKa = 11.84KK214 pKa = 9.11RR215 pKa = 11.84SCEE218 pKa = 3.93TTQPGAKK225 pKa = 8.73TNCKK229 pKa = 9.24RR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84STRR236 pKa = 11.84RR237 pKa = 3.09
Molecular weight: 26.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1746 |
169 |
634 |
349.2 |
39.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.446 ± 1.494 | 2.577 ± 0.698 |
4.926 ± 0.732 | 6.529 ± 0.591 |
3.608 ± 0.475 | 6.987 ± 0.591 |
1.775 ± 0.366 | 4.811 ± 0.883 |
5.785 ± 0.975 | 10.481 ± 0.678 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.577 ± 0.391 | 5.04 ± 0.406 |
5.212 ± 0.665 | 3.952 ± 0.365 |
5.556 ± 1.184 | 5.727 ± 0.22 |
7.045 ± 0.515 | 5.498 ± 0.548 |
1.718 ± 0.395 | 2.749 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
