
Giant panda associated partiti-like virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.59
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
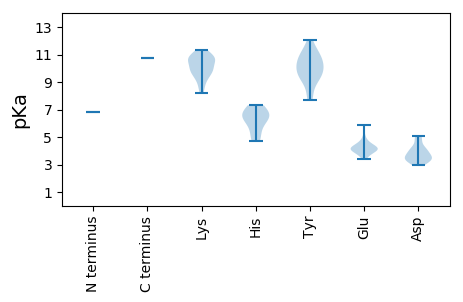
Protein with the lowest isoelectric point:
>tr|A0A220IGT0|A0A220IGT0_9VIRU RdRp OS=Giant panda associated partiti-like virus OX=2016462 PE=4 SV=1
MM1 pKa = 6.82EE2 pKa = 5.88HH3 pKa = 7.31SMKK6 pKa = 9.58TWHH9 pKa = 6.91KK10 pKa = 10.5NLDD13 pKa = 3.83KK14 pKa = 10.92EE15 pKa = 4.71SSRR18 pKa = 11.84EE19 pKa = 3.72DD20 pKa = 3.47DD21 pKa = 4.13YY22 pKa = 12.05QPVIYY27 pKa = 9.28TLSRR31 pKa = 11.84EE32 pKa = 4.1IVRR35 pKa = 11.84EE36 pKa = 3.6RR37 pKa = 11.84STQLNLNQEE46 pKa = 4.34VTSLQEE52 pKa = 3.91GPYY55 pKa = 8.12MWKK58 pKa = 8.2TVPYY62 pKa = 9.4KK63 pKa = 10.58LAALRR68 pKa = 11.84EE69 pKa = 4.27WNLRR73 pKa = 11.84MSRR76 pKa = 11.84SEE78 pKa = 3.88TPEE81 pKa = 3.43IFDD84 pKa = 3.63PVVAHH89 pKa = 6.46VLRR92 pKa = 11.84EE93 pKa = 3.82QGFEE97 pKa = 3.99VDD99 pKa = 3.13SKK101 pKa = 11.35KK102 pKa = 10.77YY103 pKa = 8.9YY104 pKa = 9.96EE105 pKa = 4.19PRR107 pKa = 11.84SVYY110 pKa = 10.35SVEE113 pKa = 4.55KK114 pKa = 10.59FFEE117 pKa = 4.15QLQSFNPSKK126 pKa = 11.0SVFVDD131 pKa = 3.35VKK133 pKa = 10.75DD134 pKa = 3.7RR135 pKa = 11.84HH136 pKa = 4.72VQRR139 pKa = 11.84GIAFAYY145 pKa = 9.72KK146 pKa = 10.45VFGKK150 pKa = 10.82GKK152 pKa = 9.93DD153 pKa = 3.51DD154 pKa = 4.75AYY156 pKa = 11.34LEE158 pKa = 4.24PLEE161 pKa = 5.09LWDD164 pKa = 5.1EE165 pKa = 4.54SIISNWKK172 pKa = 9.1GSAGVTAYY180 pKa = 10.4GQTKK184 pKa = 9.8RR185 pKa = 11.84EE186 pKa = 4.05AFSRR190 pKa = 11.84GILSAKK196 pKa = 10.0RR197 pKa = 11.84ILGQDD202 pKa = 2.97GLARR206 pKa = 11.84RR207 pKa = 11.84KK208 pKa = 9.78PEE210 pKa = 3.94PCLGLTRR217 pKa = 11.84TGKK220 pKa = 9.93NGKK223 pKa = 9.11SRR225 pKa = 11.84LIWGYY230 pKa = 9.24PMSMTLIEE238 pKa = 4.64GSLAKK243 pKa = 10.1PLLGTLTGLNTPMAYY258 pKa = 9.92KK259 pKa = 9.42LTNKK263 pKa = 8.48TLGMKK268 pKa = 9.9ILAASQTKK276 pKa = 8.73KK277 pKa = 9.36WYY279 pKa = 9.49YY280 pKa = 11.03SLDD283 pKa = 3.22ATQFDD288 pKa = 4.22SSIQKK293 pKa = 10.16IMIEE297 pKa = 4.09VAFSIIATHH306 pKa = 6.55FDD308 pKa = 3.35RR309 pKa = 11.84KK310 pKa = 10.83AMYY313 pKa = 10.66DD314 pKa = 3.45DD315 pKa = 4.14VNTVGEE321 pKa = 4.18VLEE324 pKa = 4.18LVRR327 pKa = 11.84DD328 pKa = 4.05YY329 pKa = 11.04FIHH332 pKa = 6.16TPIVMPAGEE341 pKa = 4.18KK342 pKa = 10.75AKK344 pKa = 11.14GNVQGVLYY352 pKa = 9.86YY353 pKa = 10.46GKK355 pKa = 10.15KK356 pKa = 9.76HH357 pKa = 5.35GVPSGSYY364 pKa = 7.69FTQLVDD370 pKa = 3.99SIVNIIVLGTLASKK384 pKa = 10.71FGFYY388 pKa = 10.42LSKK391 pKa = 11.36EE392 pKa = 4.3EE393 pKa = 4.26IFVLGDD399 pKa = 3.44DD400 pKa = 4.95LLFFTNTHH408 pKa = 6.57LDD410 pKa = 3.35VKK412 pKa = 10.74KK413 pKa = 10.48PSTSYY418 pKa = 11.18AKK420 pKa = 9.49STFGMLLKK428 pKa = 10.78
MM1 pKa = 6.82EE2 pKa = 5.88HH3 pKa = 7.31SMKK6 pKa = 9.58TWHH9 pKa = 6.91KK10 pKa = 10.5NLDD13 pKa = 3.83KK14 pKa = 10.92EE15 pKa = 4.71SSRR18 pKa = 11.84EE19 pKa = 3.72DD20 pKa = 3.47DD21 pKa = 4.13YY22 pKa = 12.05QPVIYY27 pKa = 9.28TLSRR31 pKa = 11.84EE32 pKa = 4.1IVRR35 pKa = 11.84EE36 pKa = 3.6RR37 pKa = 11.84STQLNLNQEE46 pKa = 4.34VTSLQEE52 pKa = 3.91GPYY55 pKa = 8.12MWKK58 pKa = 8.2TVPYY62 pKa = 9.4KK63 pKa = 10.58LAALRR68 pKa = 11.84EE69 pKa = 4.27WNLRR73 pKa = 11.84MSRR76 pKa = 11.84SEE78 pKa = 3.88TPEE81 pKa = 3.43IFDD84 pKa = 3.63PVVAHH89 pKa = 6.46VLRR92 pKa = 11.84EE93 pKa = 3.82QGFEE97 pKa = 3.99VDD99 pKa = 3.13SKK101 pKa = 11.35KK102 pKa = 10.77YY103 pKa = 8.9YY104 pKa = 9.96EE105 pKa = 4.19PRR107 pKa = 11.84SVYY110 pKa = 10.35SVEE113 pKa = 4.55KK114 pKa = 10.59FFEE117 pKa = 4.15QLQSFNPSKK126 pKa = 11.0SVFVDD131 pKa = 3.35VKK133 pKa = 10.75DD134 pKa = 3.7RR135 pKa = 11.84HH136 pKa = 4.72VQRR139 pKa = 11.84GIAFAYY145 pKa = 9.72KK146 pKa = 10.45VFGKK150 pKa = 10.82GKK152 pKa = 9.93DD153 pKa = 3.51DD154 pKa = 4.75AYY156 pKa = 11.34LEE158 pKa = 4.24PLEE161 pKa = 5.09LWDD164 pKa = 5.1EE165 pKa = 4.54SIISNWKK172 pKa = 9.1GSAGVTAYY180 pKa = 10.4GQTKK184 pKa = 9.8RR185 pKa = 11.84EE186 pKa = 4.05AFSRR190 pKa = 11.84GILSAKK196 pKa = 10.0RR197 pKa = 11.84ILGQDD202 pKa = 2.97GLARR206 pKa = 11.84RR207 pKa = 11.84KK208 pKa = 9.78PEE210 pKa = 3.94PCLGLTRR217 pKa = 11.84TGKK220 pKa = 9.93NGKK223 pKa = 9.11SRR225 pKa = 11.84LIWGYY230 pKa = 9.24PMSMTLIEE238 pKa = 4.64GSLAKK243 pKa = 10.1PLLGTLTGLNTPMAYY258 pKa = 9.92KK259 pKa = 9.42LTNKK263 pKa = 8.48TLGMKK268 pKa = 9.9ILAASQTKK276 pKa = 8.73KK277 pKa = 9.36WYY279 pKa = 9.49YY280 pKa = 11.03SLDD283 pKa = 3.22ATQFDD288 pKa = 4.22SSIQKK293 pKa = 10.16IMIEE297 pKa = 4.09VAFSIIATHH306 pKa = 6.55FDD308 pKa = 3.35RR309 pKa = 11.84KK310 pKa = 10.83AMYY313 pKa = 10.66DD314 pKa = 3.45DD315 pKa = 4.14VNTVGEE321 pKa = 4.18VLEE324 pKa = 4.18LVRR327 pKa = 11.84DD328 pKa = 4.05YY329 pKa = 11.04FIHH332 pKa = 6.16TPIVMPAGEE341 pKa = 4.18KK342 pKa = 10.75AKK344 pKa = 11.14GNVQGVLYY352 pKa = 9.86YY353 pKa = 10.46GKK355 pKa = 10.15KK356 pKa = 9.76HH357 pKa = 5.35GVPSGSYY364 pKa = 7.69FTQLVDD370 pKa = 3.99SIVNIIVLGTLASKK384 pKa = 10.71FGFYY388 pKa = 10.42LSKK391 pKa = 11.36EE392 pKa = 4.3EE393 pKa = 4.26IFVLGDD399 pKa = 3.44DD400 pKa = 4.95LLFFTNTHH408 pKa = 6.57LDD410 pKa = 3.35VKK412 pKa = 10.74KK413 pKa = 10.48PSTSYY418 pKa = 11.18AKK420 pKa = 9.49STFGMLLKK428 pKa = 10.78
Molecular weight: 48.63 kDa
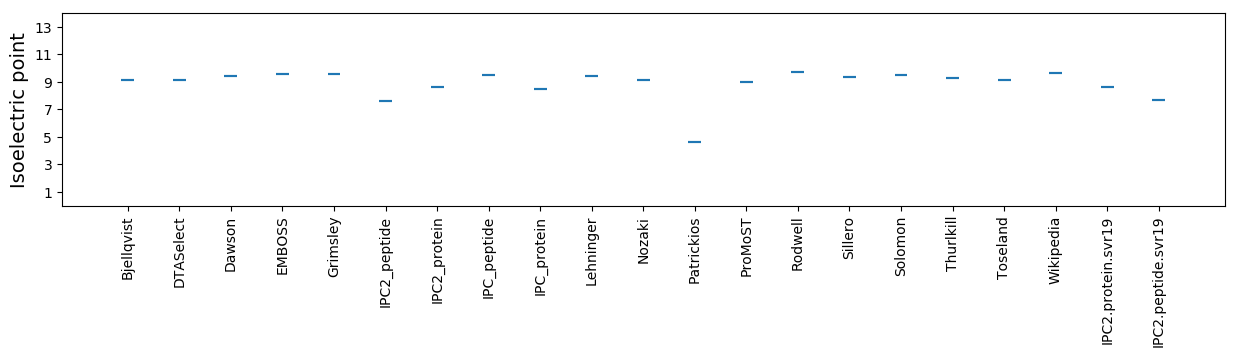
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IGT0|A0A220IGT0_9VIRU RdRp OS=Giant panda associated partiti-like virus OX=2016462 PE=4 SV=1
MM1 pKa = 6.82EE2 pKa = 5.88HH3 pKa = 7.31SMKK6 pKa = 9.58TWHH9 pKa = 6.91KK10 pKa = 10.5NLDD13 pKa = 3.83KK14 pKa = 10.92EE15 pKa = 4.71SSRR18 pKa = 11.84EE19 pKa = 3.72DD20 pKa = 3.47DD21 pKa = 4.13YY22 pKa = 12.05QPVIYY27 pKa = 9.28TLSRR31 pKa = 11.84EE32 pKa = 4.1IVRR35 pKa = 11.84EE36 pKa = 3.6RR37 pKa = 11.84STQLNLNQEE46 pKa = 4.34VTSLQEE52 pKa = 3.91GPYY55 pKa = 8.12MWKK58 pKa = 8.2TVPYY62 pKa = 9.4KK63 pKa = 10.58LAALRR68 pKa = 11.84EE69 pKa = 4.27WNLRR73 pKa = 11.84MSRR76 pKa = 11.84SEE78 pKa = 3.88TPEE81 pKa = 3.43IFDD84 pKa = 3.63PVVAHH89 pKa = 6.46VLRR92 pKa = 11.84EE93 pKa = 3.82QGFEE97 pKa = 3.99VDD99 pKa = 3.13SKK101 pKa = 11.35KK102 pKa = 10.77YY103 pKa = 8.9YY104 pKa = 9.96EE105 pKa = 4.19PRR107 pKa = 11.84SVYY110 pKa = 10.35SVEE113 pKa = 4.55KK114 pKa = 10.59FFEE117 pKa = 4.15QLQSFNPSKK126 pKa = 11.0SVFVDD131 pKa = 3.35VKK133 pKa = 10.75DD134 pKa = 3.7RR135 pKa = 11.84HH136 pKa = 4.72VQRR139 pKa = 11.84GIAFAYY145 pKa = 9.72KK146 pKa = 10.45VFGKK150 pKa = 10.82GKK152 pKa = 9.93DD153 pKa = 3.51DD154 pKa = 4.75AYY156 pKa = 11.34LEE158 pKa = 4.24PLEE161 pKa = 5.09LWDD164 pKa = 5.1EE165 pKa = 4.54SIISNWKK172 pKa = 9.1GSAGVTAYY180 pKa = 10.4GQTKK184 pKa = 9.8RR185 pKa = 11.84EE186 pKa = 4.05AFSRR190 pKa = 11.84GILSAKK196 pKa = 10.0RR197 pKa = 11.84ILGQDD202 pKa = 2.97GLARR206 pKa = 11.84RR207 pKa = 11.84KK208 pKa = 9.78PEE210 pKa = 3.94PCLGLTRR217 pKa = 11.84TGKK220 pKa = 9.93NGKK223 pKa = 9.11SRR225 pKa = 11.84LIWGYY230 pKa = 9.24PMSMTLIEE238 pKa = 4.64GSLAKK243 pKa = 10.1PLLGTLTGLNTPMAYY258 pKa = 9.92KK259 pKa = 9.42LTNKK263 pKa = 8.48TLGMKK268 pKa = 9.9ILAASQTKK276 pKa = 8.73KK277 pKa = 9.36WYY279 pKa = 9.49YY280 pKa = 11.03SLDD283 pKa = 3.22ATQFDD288 pKa = 4.22SSIQKK293 pKa = 10.16IMIEE297 pKa = 4.09VAFSIIATHH306 pKa = 6.55FDD308 pKa = 3.35RR309 pKa = 11.84KK310 pKa = 10.83AMYY313 pKa = 10.66DD314 pKa = 3.45DD315 pKa = 4.14VNTVGEE321 pKa = 4.18VLEE324 pKa = 4.18LVRR327 pKa = 11.84DD328 pKa = 4.05YY329 pKa = 11.04FIHH332 pKa = 6.16TPIVMPAGEE341 pKa = 4.18KK342 pKa = 10.75AKK344 pKa = 11.14GNVQGVLYY352 pKa = 9.86YY353 pKa = 10.46GKK355 pKa = 10.15KK356 pKa = 9.76HH357 pKa = 5.35GVPSGSYY364 pKa = 7.69FTQLVDD370 pKa = 3.99SIVNIIVLGTLASKK384 pKa = 10.71FGFYY388 pKa = 10.42LSKK391 pKa = 11.36EE392 pKa = 4.3EE393 pKa = 4.26IFVLGDD399 pKa = 3.44DD400 pKa = 4.95LLFFTNTHH408 pKa = 6.57LDD410 pKa = 3.35VKK412 pKa = 10.74KK413 pKa = 10.48PSTSYY418 pKa = 11.18AKK420 pKa = 9.49STFGMLLKK428 pKa = 10.78
MM1 pKa = 6.82EE2 pKa = 5.88HH3 pKa = 7.31SMKK6 pKa = 9.58TWHH9 pKa = 6.91KK10 pKa = 10.5NLDD13 pKa = 3.83KK14 pKa = 10.92EE15 pKa = 4.71SSRR18 pKa = 11.84EE19 pKa = 3.72DD20 pKa = 3.47DD21 pKa = 4.13YY22 pKa = 12.05QPVIYY27 pKa = 9.28TLSRR31 pKa = 11.84EE32 pKa = 4.1IVRR35 pKa = 11.84EE36 pKa = 3.6RR37 pKa = 11.84STQLNLNQEE46 pKa = 4.34VTSLQEE52 pKa = 3.91GPYY55 pKa = 8.12MWKK58 pKa = 8.2TVPYY62 pKa = 9.4KK63 pKa = 10.58LAALRR68 pKa = 11.84EE69 pKa = 4.27WNLRR73 pKa = 11.84MSRR76 pKa = 11.84SEE78 pKa = 3.88TPEE81 pKa = 3.43IFDD84 pKa = 3.63PVVAHH89 pKa = 6.46VLRR92 pKa = 11.84EE93 pKa = 3.82QGFEE97 pKa = 3.99VDD99 pKa = 3.13SKK101 pKa = 11.35KK102 pKa = 10.77YY103 pKa = 8.9YY104 pKa = 9.96EE105 pKa = 4.19PRR107 pKa = 11.84SVYY110 pKa = 10.35SVEE113 pKa = 4.55KK114 pKa = 10.59FFEE117 pKa = 4.15QLQSFNPSKK126 pKa = 11.0SVFVDD131 pKa = 3.35VKK133 pKa = 10.75DD134 pKa = 3.7RR135 pKa = 11.84HH136 pKa = 4.72VQRR139 pKa = 11.84GIAFAYY145 pKa = 9.72KK146 pKa = 10.45VFGKK150 pKa = 10.82GKK152 pKa = 9.93DD153 pKa = 3.51DD154 pKa = 4.75AYY156 pKa = 11.34LEE158 pKa = 4.24PLEE161 pKa = 5.09LWDD164 pKa = 5.1EE165 pKa = 4.54SIISNWKK172 pKa = 9.1GSAGVTAYY180 pKa = 10.4GQTKK184 pKa = 9.8RR185 pKa = 11.84EE186 pKa = 4.05AFSRR190 pKa = 11.84GILSAKK196 pKa = 10.0RR197 pKa = 11.84ILGQDD202 pKa = 2.97GLARR206 pKa = 11.84RR207 pKa = 11.84KK208 pKa = 9.78PEE210 pKa = 3.94PCLGLTRR217 pKa = 11.84TGKK220 pKa = 9.93NGKK223 pKa = 9.11SRR225 pKa = 11.84LIWGYY230 pKa = 9.24PMSMTLIEE238 pKa = 4.64GSLAKK243 pKa = 10.1PLLGTLTGLNTPMAYY258 pKa = 9.92KK259 pKa = 9.42LTNKK263 pKa = 8.48TLGMKK268 pKa = 9.9ILAASQTKK276 pKa = 8.73KK277 pKa = 9.36WYY279 pKa = 9.49YY280 pKa = 11.03SLDD283 pKa = 3.22ATQFDD288 pKa = 4.22SSIQKK293 pKa = 10.16IMIEE297 pKa = 4.09VAFSIIATHH306 pKa = 6.55FDD308 pKa = 3.35RR309 pKa = 11.84KK310 pKa = 10.83AMYY313 pKa = 10.66DD314 pKa = 3.45DD315 pKa = 4.14VNTVGEE321 pKa = 4.18VLEE324 pKa = 4.18LVRR327 pKa = 11.84DD328 pKa = 4.05YY329 pKa = 11.04FIHH332 pKa = 6.16TPIVMPAGEE341 pKa = 4.18KK342 pKa = 10.75AKK344 pKa = 11.14GNVQGVLYY352 pKa = 9.86YY353 pKa = 10.46GKK355 pKa = 10.15KK356 pKa = 9.76HH357 pKa = 5.35GVPSGSYY364 pKa = 7.69FTQLVDD370 pKa = 3.99SIVNIIVLGTLASKK384 pKa = 10.71FGFYY388 pKa = 10.42LSKK391 pKa = 11.36EE392 pKa = 4.3EE393 pKa = 4.26IFVLGDD399 pKa = 3.44DD400 pKa = 4.95LLFFTNTHH408 pKa = 6.57LDD410 pKa = 3.35VKK412 pKa = 10.74KK413 pKa = 10.48PSTSYY418 pKa = 11.18AKK420 pKa = 9.49STFGMLLKK428 pKa = 10.78
Molecular weight: 48.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
428 |
428 |
428 |
428.0 |
48.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.374 ± 0.0 | 0.234 ± 0.0 |
4.907 ± 0.0 | 6.308 ± 0.0 |
4.673 ± 0.0 | 7.009 ± 0.0 |
1.869 ± 0.0 | 5.14 ± 0.0 |
8.645 ± 0.0 | 9.813 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.804 ± 0.0 | 3.037 ± 0.0 |
3.972 ± 0.0 | 3.505 ± 0.0 |
4.673 ± 0.0 | 8.178 ± 0.0 |
6.308 ± 0.0 | 7.009 ± 0.0 |
1.636 ± 0.0 | 4.907 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
