
Thiorhodococcus drewsii AZ1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Thiorhodococcus; Thiorhodococcus drewsii
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
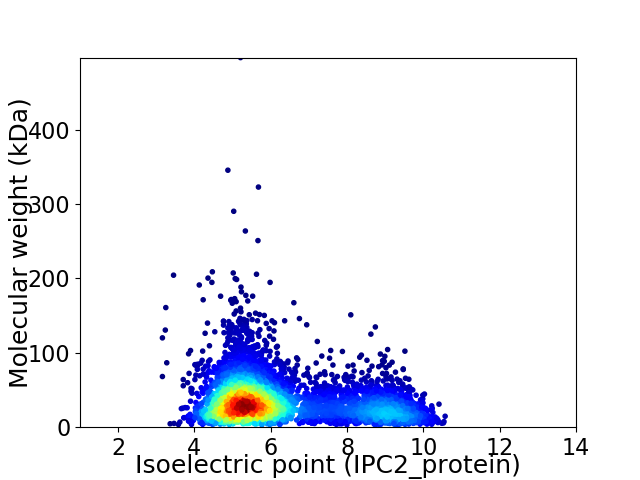
Virtual 2D-PAGE plot for 4731 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2E7E9|G2E7E9_9GAMM Methyltransferase type 11 OS=Thiorhodococcus drewsii AZ1 OX=765913 GN=ThidrDRAFT_4212 PE=4 SV=1
MM1 pKa = 7.94IIYY4 pKa = 9.54TDD6 pKa = 4.37LAQPLLDD13 pKa = 3.41YY14 pKa = 11.1VSAVEE19 pKa = 4.68AEE21 pKa = 4.13ASGTSDD27 pKa = 3.77SLNVDD32 pKa = 4.0LTLSADD38 pKa = 4.43FFAKK42 pKa = 10.31RR43 pKa = 11.84PQILTANFGFDD54 pKa = 3.18GYY56 pKa = 10.17MGVPGLVGADD66 pKa = 3.23ATAQSAAYY74 pKa = 8.63QFNVAWEE81 pKa = 4.28ALDD84 pKa = 3.99PALGAATRR92 pKa = 11.84AYY94 pKa = 10.07YY95 pKa = 10.54SAVGPAAFLSVYY107 pKa = 9.78GVPALLADD115 pKa = 4.87TIPVVFSHH123 pKa = 6.88PVLGEE128 pKa = 4.01TVNPEE133 pKa = 4.05DD134 pKa = 5.25FLVTLNTGEE143 pKa = 4.24VVTPITASFLPNLEE157 pKa = 3.99FNEE160 pKa = 4.14RR161 pKa = 11.84QTVVISGEE169 pKa = 3.75WGNRR173 pKa = 11.84IAPGEE178 pKa = 4.07PGARR182 pKa = 11.84YY183 pKa = 8.21PVTVSIVDD191 pKa = 4.05DD192 pKa = 4.59GSPLCFVTDD201 pKa = 3.44TGLISAVGLSVEE213 pKa = 4.28SQNPYY218 pKa = 10.27VEE220 pKa = 4.87GNGPRR225 pKa = 11.84ILAAKK230 pKa = 10.11LNAFSDD236 pKa = 3.91LGEE239 pKa = 5.08GSPAWQGSSTANSGADD255 pKa = 3.51LYY257 pKa = 11.38GDD259 pKa = 3.17QAMYY263 pKa = 10.8RR264 pKa = 11.84LRR266 pKa = 11.84IYY268 pKa = 10.14TSAGFSPDD276 pKa = 3.58GIASISPDD284 pKa = 3.18DD285 pKa = 3.85FSRR288 pKa = 11.84FFQLTALDD296 pKa = 3.85QGGHH300 pKa = 5.62EE301 pKa = 4.69LLLLEE306 pKa = 4.57TGVDD310 pKa = 3.94YY311 pKa = 11.35VISGFGAVRR320 pKa = 11.84ILGIADD326 pKa = 3.4TGLVQDD332 pKa = 5.13SYY334 pKa = 10.82NAAYY338 pKa = 9.7IEE340 pKa = 4.38DD341 pKa = 4.6HH342 pKa = 7.25DD343 pKa = 4.12NQYY346 pKa = 11.49DD347 pKa = 3.84IILSGDD353 pKa = 3.32LAAIEE358 pKa = 4.36RR359 pKa = 11.84LQSVRR364 pKa = 11.84MPSGDD369 pKa = 3.34GYY371 pKa = 11.39SPVYY375 pKa = 10.75NPGGPGNDD383 pKa = 4.03PDD385 pKa = 4.55NNPPGPFTVPSSDD398 pKa = 3.15QTVDD402 pKa = 2.7ITHH405 pKa = 7.6DD406 pKa = 3.54FTAASFVSYY415 pKa = 11.08VEE417 pKa = 3.99VDD419 pKa = 3.61GQVVHH424 pKa = 6.55HH425 pKa = 7.55AITGQPIGVDD435 pKa = 2.86QGLAVEE441 pKa = 4.66DD442 pKa = 4.17TSTGHH447 pKa = 7.59KK448 pKa = 9.26INQYY452 pKa = 9.95IDD454 pKa = 3.39PDD456 pKa = 3.76GKK458 pKa = 10.09IFYY461 pKa = 10.46SSFEE465 pKa = 3.69VSPIYY470 pKa = 9.6EE471 pKa = 3.69IHH473 pKa = 5.83LTEE476 pKa = 4.4VQPSTYY482 pKa = 10.75SRR484 pKa = 11.84VTDD487 pKa = 3.84DD488 pKa = 4.47QITGSAQIDD497 pKa = 3.29RR498 pKa = 11.84VSFSGAFDD506 pKa = 3.24QYY508 pKa = 11.24VRR510 pKa = 11.84SGNLAEE516 pKa = 5.71FSSQDD521 pKa = 3.69LVPLRR526 pKa = 11.84DD527 pKa = 3.56ADD529 pKa = 4.65DD530 pKa = 3.83NLFDD534 pKa = 4.14VEE536 pKa = 4.24RR537 pKa = 11.84LIYY540 pKa = 10.47TDD542 pKa = 3.14TTLALDD548 pKa = 3.91IEE550 pKa = 4.74GHH552 pKa = 6.48AGFAYY557 pKa = 10.3SLYY560 pKa = 10.54GVFGRR565 pKa = 11.84APDD568 pKa = 3.66AAGLGDD574 pKa = 5.07WINALDD580 pKa = 4.26NGFSLQSVADD590 pKa = 4.21RR591 pKa = 11.84FLASPEE597 pKa = 4.17FKK599 pKa = 9.29THH601 pKa = 6.23YY602 pKa = 10.37GPPLTNGSYY611 pKa = 10.44VEE613 pKa = 3.79QLYY616 pKa = 11.12EE617 pKa = 3.93NFLNRR622 pKa = 11.84SAEE625 pKa = 3.97ASGYY629 pKa = 8.86DD630 pKa = 3.35AWVGALDD637 pKa = 4.7AGLLDD642 pKa = 4.36RR643 pKa = 11.84SDD645 pKa = 3.65VLIAFSRR652 pKa = 11.84SPEE655 pKa = 4.01YY656 pKa = 10.4QDD658 pKa = 6.05LIAGDD663 pKa = 3.64IANGIRR669 pKa = 11.84FDD671 pKa = 4.52LWWQQ675 pKa = 3.28
MM1 pKa = 7.94IIYY4 pKa = 9.54TDD6 pKa = 4.37LAQPLLDD13 pKa = 3.41YY14 pKa = 11.1VSAVEE19 pKa = 4.68AEE21 pKa = 4.13ASGTSDD27 pKa = 3.77SLNVDD32 pKa = 4.0LTLSADD38 pKa = 4.43FFAKK42 pKa = 10.31RR43 pKa = 11.84PQILTANFGFDD54 pKa = 3.18GYY56 pKa = 10.17MGVPGLVGADD66 pKa = 3.23ATAQSAAYY74 pKa = 8.63QFNVAWEE81 pKa = 4.28ALDD84 pKa = 3.99PALGAATRR92 pKa = 11.84AYY94 pKa = 10.07YY95 pKa = 10.54SAVGPAAFLSVYY107 pKa = 9.78GVPALLADD115 pKa = 4.87TIPVVFSHH123 pKa = 6.88PVLGEE128 pKa = 4.01TVNPEE133 pKa = 4.05DD134 pKa = 5.25FLVTLNTGEE143 pKa = 4.24VVTPITASFLPNLEE157 pKa = 3.99FNEE160 pKa = 4.14RR161 pKa = 11.84QTVVISGEE169 pKa = 3.75WGNRR173 pKa = 11.84IAPGEE178 pKa = 4.07PGARR182 pKa = 11.84YY183 pKa = 8.21PVTVSIVDD191 pKa = 4.05DD192 pKa = 4.59GSPLCFVTDD201 pKa = 3.44TGLISAVGLSVEE213 pKa = 4.28SQNPYY218 pKa = 10.27VEE220 pKa = 4.87GNGPRR225 pKa = 11.84ILAAKK230 pKa = 10.11LNAFSDD236 pKa = 3.91LGEE239 pKa = 5.08GSPAWQGSSTANSGADD255 pKa = 3.51LYY257 pKa = 11.38GDD259 pKa = 3.17QAMYY263 pKa = 10.8RR264 pKa = 11.84LRR266 pKa = 11.84IYY268 pKa = 10.14TSAGFSPDD276 pKa = 3.58GIASISPDD284 pKa = 3.18DD285 pKa = 3.85FSRR288 pKa = 11.84FFQLTALDD296 pKa = 3.85QGGHH300 pKa = 5.62EE301 pKa = 4.69LLLLEE306 pKa = 4.57TGVDD310 pKa = 3.94YY311 pKa = 11.35VISGFGAVRR320 pKa = 11.84ILGIADD326 pKa = 3.4TGLVQDD332 pKa = 5.13SYY334 pKa = 10.82NAAYY338 pKa = 9.7IEE340 pKa = 4.38DD341 pKa = 4.6HH342 pKa = 7.25DD343 pKa = 4.12NQYY346 pKa = 11.49DD347 pKa = 3.84IILSGDD353 pKa = 3.32LAAIEE358 pKa = 4.36RR359 pKa = 11.84LQSVRR364 pKa = 11.84MPSGDD369 pKa = 3.34GYY371 pKa = 11.39SPVYY375 pKa = 10.75NPGGPGNDD383 pKa = 4.03PDD385 pKa = 4.55NNPPGPFTVPSSDD398 pKa = 3.15QTVDD402 pKa = 2.7ITHH405 pKa = 7.6DD406 pKa = 3.54FTAASFVSYY415 pKa = 11.08VEE417 pKa = 3.99VDD419 pKa = 3.61GQVVHH424 pKa = 6.55HH425 pKa = 7.55AITGQPIGVDD435 pKa = 2.86QGLAVEE441 pKa = 4.66DD442 pKa = 4.17TSTGHH447 pKa = 7.59KK448 pKa = 9.26INQYY452 pKa = 9.95IDD454 pKa = 3.39PDD456 pKa = 3.76GKK458 pKa = 10.09IFYY461 pKa = 10.46SSFEE465 pKa = 3.69VSPIYY470 pKa = 9.6EE471 pKa = 3.69IHH473 pKa = 5.83LTEE476 pKa = 4.4VQPSTYY482 pKa = 10.75SRR484 pKa = 11.84VTDD487 pKa = 3.84DD488 pKa = 4.47QITGSAQIDD497 pKa = 3.29RR498 pKa = 11.84VSFSGAFDD506 pKa = 3.24QYY508 pKa = 11.24VRR510 pKa = 11.84SGNLAEE516 pKa = 5.71FSSQDD521 pKa = 3.69LVPLRR526 pKa = 11.84DD527 pKa = 3.56ADD529 pKa = 4.65DD530 pKa = 3.83NLFDD534 pKa = 4.14VEE536 pKa = 4.24RR537 pKa = 11.84LIYY540 pKa = 10.47TDD542 pKa = 3.14TTLALDD548 pKa = 3.91IEE550 pKa = 4.74GHH552 pKa = 6.48AGFAYY557 pKa = 10.3SLYY560 pKa = 10.54GVFGRR565 pKa = 11.84APDD568 pKa = 3.66AAGLGDD574 pKa = 5.07WINALDD580 pKa = 4.26NGFSLQSVADD590 pKa = 4.21RR591 pKa = 11.84FLASPEE597 pKa = 4.17FKK599 pKa = 9.29THH601 pKa = 6.23YY602 pKa = 10.37GPPLTNGSYY611 pKa = 10.44VEE613 pKa = 3.79QLYY616 pKa = 11.12EE617 pKa = 3.93NFLNRR622 pKa = 11.84SAEE625 pKa = 3.97ASGYY629 pKa = 8.86DD630 pKa = 3.35AWVGALDD637 pKa = 4.7AGLLDD642 pKa = 4.36RR643 pKa = 11.84SDD645 pKa = 3.65VLIAFSRR652 pKa = 11.84SPEE655 pKa = 4.01YY656 pKa = 10.4QDD658 pKa = 6.05LIAGDD663 pKa = 3.64IANGIRR669 pKa = 11.84FDD671 pKa = 4.52LWWQQ675 pKa = 3.28
Molecular weight: 72.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2DXC3|G2DXC3_9GAMM Uncharacterized protein OS=Thiorhodococcus drewsii AZ1 OX=765913 GN=ThidrDRAFT_0684 PE=4 SV=1
MM1 pKa = 7.36RR2 pKa = 11.84QVPLEE7 pKa = 4.26SNSARR12 pKa = 11.84EE13 pKa = 4.22PKK15 pKa = 10.39VGAGVVGRR23 pKa = 11.84SVGGLRR29 pKa = 11.84LFASCASFVLLCLVLARR46 pKa = 11.84SHH48 pKa = 6.66AEE50 pKa = 3.73SFSEE54 pKa = 4.33VEE56 pKa = 3.93VRR58 pKa = 11.84AVYY61 pKa = 10.38LFNFAVFVDD70 pKa = 4.33WPPTAFEE77 pKa = 4.88SSTSPIRR84 pKa = 11.84YY85 pKa = 8.82CVVGSQALRR94 pKa = 11.84ASLEE98 pKa = 3.95EE99 pKa = 4.09VLADD103 pKa = 3.38EE104 pKa = 4.74HH105 pKa = 7.66VGGRR109 pKa = 11.84ALRR112 pKa = 11.84LISANEE118 pKa = 3.61PLDD121 pKa = 3.12WRR123 pKa = 11.84RR124 pKa = 11.84CHH126 pKa = 6.98LLYY129 pKa = 10.26IGHH132 pKa = 6.39GAKK135 pKa = 9.0TRR137 pKa = 11.84ARR139 pKa = 11.84QILKK143 pKa = 10.06AVRR146 pKa = 11.84GTAVLTIGASEE157 pKa = 4.11AFVRR161 pKa = 11.84EE162 pKa = 3.85GGMIGLVRR170 pKa = 11.84RR171 pKa = 11.84GQRR174 pKa = 11.84LRR176 pKa = 11.84TIIDD180 pKa = 2.89RR181 pKa = 11.84DD182 pKa = 3.15AAEE185 pKa = 3.94RR186 pKa = 11.84VGIRR190 pKa = 11.84ISSKK194 pKa = 10.24LLRR197 pKa = 11.84LSTLVSGTRR206 pKa = 11.84EE207 pKa = 4.13GVSPP211 pKa = 4.5
MM1 pKa = 7.36RR2 pKa = 11.84QVPLEE7 pKa = 4.26SNSARR12 pKa = 11.84EE13 pKa = 4.22PKK15 pKa = 10.39VGAGVVGRR23 pKa = 11.84SVGGLRR29 pKa = 11.84LFASCASFVLLCLVLARR46 pKa = 11.84SHH48 pKa = 6.66AEE50 pKa = 3.73SFSEE54 pKa = 4.33VEE56 pKa = 3.93VRR58 pKa = 11.84AVYY61 pKa = 10.38LFNFAVFVDD70 pKa = 4.33WPPTAFEE77 pKa = 4.88SSTSPIRR84 pKa = 11.84YY85 pKa = 8.82CVVGSQALRR94 pKa = 11.84ASLEE98 pKa = 3.95EE99 pKa = 4.09VLADD103 pKa = 3.38EE104 pKa = 4.74HH105 pKa = 7.66VGGRR109 pKa = 11.84ALRR112 pKa = 11.84LISANEE118 pKa = 3.61PLDD121 pKa = 3.12WRR123 pKa = 11.84RR124 pKa = 11.84CHH126 pKa = 6.98LLYY129 pKa = 10.26IGHH132 pKa = 6.39GAKK135 pKa = 9.0TRR137 pKa = 11.84ARR139 pKa = 11.84QILKK143 pKa = 10.06AVRR146 pKa = 11.84GTAVLTIGASEE157 pKa = 4.11AFVRR161 pKa = 11.84EE162 pKa = 3.85GGMIGLVRR170 pKa = 11.84RR171 pKa = 11.84GQRR174 pKa = 11.84LRR176 pKa = 11.84TIIDD180 pKa = 2.89RR181 pKa = 11.84DD182 pKa = 3.15AAEE185 pKa = 3.94RR186 pKa = 11.84VGIRR190 pKa = 11.84ISSKK194 pKa = 10.24LLRR197 pKa = 11.84LSTLVSGTRR206 pKa = 11.84EE207 pKa = 4.13GVSPP211 pKa = 4.5
Molecular weight: 22.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1618336 |
30 |
4605 |
342.1 |
37.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.771 ± 0.041 | 1.023 ± 0.013 |
6.068 ± 0.03 | 6.345 ± 0.033 |
3.473 ± 0.02 | 7.864 ± 0.036 |
2.254 ± 0.017 | 4.992 ± 0.026 |
2.772 ± 0.025 | 11.415 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.211 ± 0.018 | 2.564 ± 0.017 |
5.128 ± 0.029 | 3.803 ± 0.025 |
7.771 ± 0.042 | 5.815 ± 0.026 |
5.094 ± 0.031 | 6.826 ± 0.03 |
1.439 ± 0.016 | 2.374 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
