
Desulfoplanes formicivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfomicrobiaceae; Desulfoplanes
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
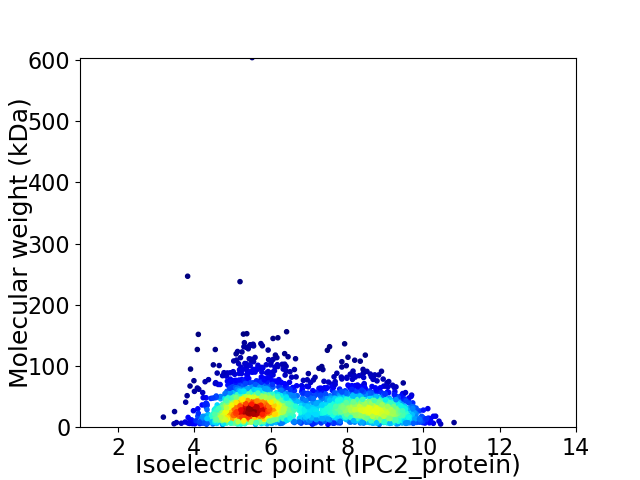
Virtual 2D-PAGE plot for 2657 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A194AG35|A0A194AG35_9DELT FMN reductase OS=Desulfoplanes formicivorans OX=1592317 GN=DPF_0437 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84VIEE5 pKa = 5.18PIDD8 pKa = 3.46IEE10 pKa = 5.51LIDD13 pKa = 4.46SNVTQSGLSEE23 pKa = 3.89WDD25 pKa = 3.17AEE27 pKa = 4.43TIYY30 pKa = 11.05AAGQRR35 pKa = 11.84VVVSYY40 pKa = 11.03EE41 pKa = 3.94EE42 pKa = 5.21DD43 pKa = 3.82GEE45 pKa = 4.56TPRR48 pKa = 11.84VAEE51 pKa = 4.81EE52 pKa = 4.05YY53 pKa = 10.22TSLRR57 pKa = 11.84SDD59 pKa = 2.94NVGFYY64 pKa = 10.54PPDD67 pKa = 3.49SSGDD71 pKa = 3.53GQDD74 pKa = 3.24GVKK77 pKa = 10.2ISVGDD82 pKa = 3.97SEE84 pKa = 5.41DD85 pKa = 3.41VSSFTIGEE93 pKa = 4.46TVGTSLASGTCSFIDD108 pKa = 4.02GTTGGYY114 pKa = 8.97MILSGVSGDD123 pKa = 3.96FDD125 pKa = 4.45SGDD128 pKa = 3.83TISGNDD134 pKa = 3.31SGASATISSVSSTTIAAAWEE154 pKa = 4.17RR155 pKa = 11.84VGATNRR161 pKa = 11.84WKK163 pKa = 10.61MFDD166 pKa = 3.78AYY168 pKa = 11.39VNSQTVADD176 pKa = 5.57DD177 pKa = 3.38IDD179 pKa = 3.77VTIGFGRR186 pKa = 11.84CDD188 pKa = 3.62TLVLLDD194 pKa = 3.77VQATDD199 pKa = 3.67VEE201 pKa = 4.53IEE203 pKa = 4.29VVNDD207 pKa = 3.29STGEE211 pKa = 4.23VVFSEE216 pKa = 4.46AFDD219 pKa = 3.62MRR221 pKa = 11.84LDD223 pKa = 4.32QSASWADD230 pKa = 3.71YY231 pKa = 9.54FFAPAEE237 pKa = 4.35YY238 pKa = 8.09KK239 pKa = 10.23TSLIVSTPLYY249 pKa = 9.28YY250 pKa = 10.39NASARR255 pKa = 11.84IRR257 pKa = 11.84ITNGDD262 pKa = 3.84DD263 pKa = 3.19EE264 pKa = 5.31AMCGHH269 pKa = 6.23VVAGRR274 pKa = 11.84AQYY277 pKa = 10.15IAASQWSPTAGITDD291 pKa = 3.72YY292 pKa = 11.56SSAEE296 pKa = 3.94TDD298 pKa = 3.18TDD300 pKa = 3.76GVTSLEE306 pKa = 3.78QGPWSKK312 pKa = 10.64EE313 pKa = 3.62LSVDD317 pKa = 3.79LLVPTASVSNLQRR330 pKa = 11.84RR331 pKa = 11.84LASYY335 pKa = 10.13RR336 pKa = 11.84GKK338 pKa = 9.96PCVWDD343 pKa = 4.1CNNDD347 pKa = 3.78STDD350 pKa = 3.17HH351 pKa = 7.16DD352 pKa = 4.01ALIVYY357 pKa = 8.86GYY359 pKa = 10.36FSDD362 pKa = 4.49FSIVIAGPTTSSCSLTIQGLII383 pKa = 3.77
MM1 pKa = 7.62RR2 pKa = 11.84VIEE5 pKa = 5.18PIDD8 pKa = 3.46IEE10 pKa = 5.51LIDD13 pKa = 4.46SNVTQSGLSEE23 pKa = 3.89WDD25 pKa = 3.17AEE27 pKa = 4.43TIYY30 pKa = 11.05AAGQRR35 pKa = 11.84VVVSYY40 pKa = 11.03EE41 pKa = 3.94EE42 pKa = 5.21DD43 pKa = 3.82GEE45 pKa = 4.56TPRR48 pKa = 11.84VAEE51 pKa = 4.81EE52 pKa = 4.05YY53 pKa = 10.22TSLRR57 pKa = 11.84SDD59 pKa = 2.94NVGFYY64 pKa = 10.54PPDD67 pKa = 3.49SSGDD71 pKa = 3.53GQDD74 pKa = 3.24GVKK77 pKa = 10.2ISVGDD82 pKa = 3.97SEE84 pKa = 5.41DD85 pKa = 3.41VSSFTIGEE93 pKa = 4.46TVGTSLASGTCSFIDD108 pKa = 4.02GTTGGYY114 pKa = 8.97MILSGVSGDD123 pKa = 3.96FDD125 pKa = 4.45SGDD128 pKa = 3.83TISGNDD134 pKa = 3.31SGASATISSVSSTTIAAAWEE154 pKa = 4.17RR155 pKa = 11.84VGATNRR161 pKa = 11.84WKK163 pKa = 10.61MFDD166 pKa = 3.78AYY168 pKa = 11.39VNSQTVADD176 pKa = 5.57DD177 pKa = 3.38IDD179 pKa = 3.77VTIGFGRR186 pKa = 11.84CDD188 pKa = 3.62TLVLLDD194 pKa = 3.77VQATDD199 pKa = 3.67VEE201 pKa = 4.53IEE203 pKa = 4.29VVNDD207 pKa = 3.29STGEE211 pKa = 4.23VVFSEE216 pKa = 4.46AFDD219 pKa = 3.62MRR221 pKa = 11.84LDD223 pKa = 4.32QSASWADD230 pKa = 3.71YY231 pKa = 9.54FFAPAEE237 pKa = 4.35YY238 pKa = 8.09KK239 pKa = 10.23TSLIVSTPLYY249 pKa = 9.28YY250 pKa = 10.39NASARR255 pKa = 11.84IRR257 pKa = 11.84ITNGDD262 pKa = 3.84DD263 pKa = 3.19EE264 pKa = 5.31AMCGHH269 pKa = 6.23VVAGRR274 pKa = 11.84AQYY277 pKa = 10.15IAASQWSPTAGITDD291 pKa = 3.72YY292 pKa = 11.56SSAEE296 pKa = 3.94TDD298 pKa = 3.18TDD300 pKa = 3.76GVTSLEE306 pKa = 3.78QGPWSKK312 pKa = 10.64EE313 pKa = 3.62LSVDD317 pKa = 3.79LLVPTASVSNLQRR330 pKa = 11.84RR331 pKa = 11.84LASYY335 pKa = 10.13RR336 pKa = 11.84GKK338 pKa = 9.96PCVWDD343 pKa = 4.1CNNDD347 pKa = 3.78STDD350 pKa = 3.17HH351 pKa = 7.16DD352 pKa = 4.01ALIVYY357 pKa = 8.86GYY359 pKa = 10.36FSDD362 pKa = 4.49FSIVIAGPTTSSCSLTIQGLII383 pKa = 3.77
Molecular weight: 40.84 kDa
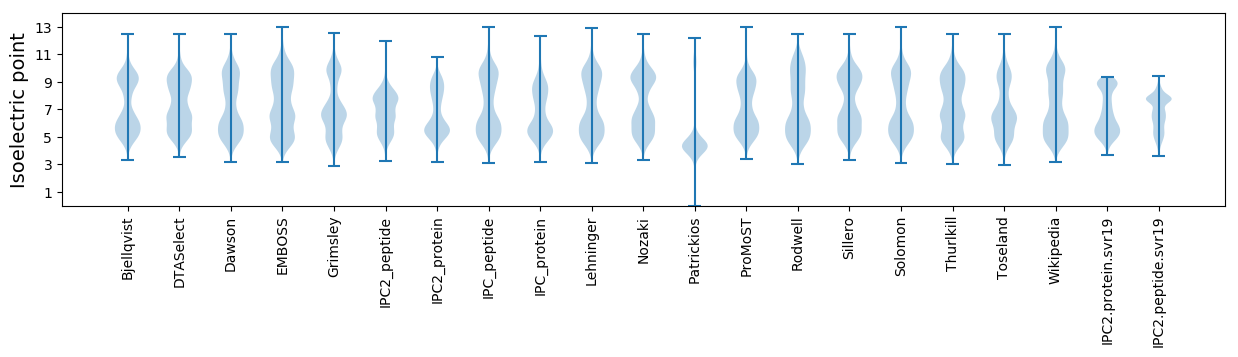
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A194AHT9|A0A194AHT9_9DELT Uncharacterized protein OS=Desulfoplanes formicivorans OX=1592317 GN=DPF_1045 PE=4 SV=1
MM1 pKa = 7.86PKK3 pKa = 10.65LKK5 pKa = 9.79TNKK8 pKa = 9.61SAAKK12 pKa = 10.15RR13 pKa = 11.84FTTTGTGKK21 pKa = 10.25VKK23 pKa = 10.65RR24 pKa = 11.84NRR26 pKa = 11.84SNSQHH31 pKa = 6.67ILTKK35 pKa = 10.63KK36 pKa = 7.69NAKK39 pKa = 9.18RR40 pKa = 11.84KK41 pKa = 9.01RR42 pKa = 11.84RR43 pKa = 11.84LRR45 pKa = 11.84QSTTMDD51 pKa = 3.01KK52 pKa = 10.13STLKK56 pKa = 10.26SVKK59 pKa = 9.98RR60 pKa = 11.84LVPTIFF66 pKa = 3.73
MM1 pKa = 7.86PKK3 pKa = 10.65LKK5 pKa = 9.79TNKK8 pKa = 9.61SAAKK12 pKa = 10.15RR13 pKa = 11.84FTTTGTGKK21 pKa = 10.25VKK23 pKa = 10.65RR24 pKa = 11.84NRR26 pKa = 11.84SNSQHH31 pKa = 6.67ILTKK35 pKa = 10.63KK36 pKa = 7.69NAKK39 pKa = 9.18RR40 pKa = 11.84KK41 pKa = 9.01RR42 pKa = 11.84RR43 pKa = 11.84LRR45 pKa = 11.84QSTTMDD51 pKa = 3.01KK52 pKa = 10.13STLKK56 pKa = 10.26SVKK59 pKa = 9.98RR60 pKa = 11.84LVPTIFF66 pKa = 3.73
Molecular weight: 7.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
859764 |
41 |
5430 |
323.6 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.662 ± 0.051 | 1.423 ± 0.021 |
5.498 ± 0.037 | 5.819 ± 0.05 |
4.164 ± 0.031 | 7.634 ± 0.039 |
2.435 ± 0.025 | 6.18 ± 0.05 |
4.932 ± 0.047 | 10.49 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.94 ± 0.02 | 3.222 ± 0.028 |
4.735 ± 0.036 | 3.85 ± 0.027 |
5.896 ± 0.037 | 5.543 ± 0.037 |
5.495 ± 0.032 | 7.122 ± 0.044 |
1.232 ± 0.02 | 2.728 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
