
Wenzhou picorna-like virus 33
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
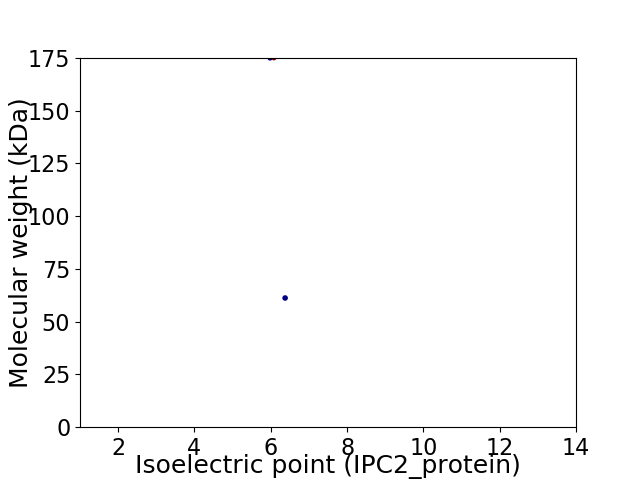
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KMD5|A0A1L3KMD5_9VIRU Uncharacterized protein OS=Wenzhou picorna-like virus 33 OX=1923619 PE=4 SV=1
MM1 pKa = 7.62SFTTNEE7 pKa = 3.99SEE9 pKa = 4.36VVVNVKK15 pKa = 9.99LLRR18 pKa = 11.84RR19 pKa = 11.84AEE21 pKa = 3.91RR22 pKa = 11.84EE23 pKa = 4.09AIQRR27 pKa = 11.84ACNIRR32 pKa = 11.84RR33 pKa = 11.84LKK35 pKa = 10.64KK36 pKa = 10.4AFKK39 pKa = 10.38SLPKK43 pKa = 10.1YY44 pKa = 9.98EE45 pKa = 4.18RR46 pKa = 11.84EE47 pKa = 3.89QVIASARR54 pKa = 11.84DD55 pKa = 3.27AKK57 pKa = 10.95YY58 pKa = 10.34SFVRR62 pKa = 11.84EE63 pKa = 3.82EE64 pKa = 4.24SGITDD69 pKa = 3.85YY70 pKa = 11.4LYY72 pKa = 11.31GAAAVAAGAAGAAAGFSIAKK92 pKa = 8.23TSHH95 pKa = 6.63EE96 pKa = 3.89IGEE99 pKa = 4.37AVKK102 pKa = 9.7DD103 pKa = 3.71TLSGVNAVMQPAGAIANSVGGALEE127 pKa = 3.93YY128 pKa = 10.56LYY130 pKa = 11.34SLVKK134 pKa = 10.11QVYY137 pKa = 7.49DD138 pKa = 3.42TCANIVGKK146 pKa = 10.05FSQLIFGIFVVAMKK160 pKa = 10.7ACFGLVNMASRR171 pKa = 11.84LFDD174 pKa = 3.53SVVSMVAPDD183 pKa = 3.58FHH185 pKa = 8.52LLDD188 pKa = 4.14SLEE191 pKa = 4.4EE192 pKa = 4.87DD193 pKa = 4.12DD194 pKa = 5.93VLPQSGCMSYY204 pKa = 10.8LPQVVTLVCTFFVPTYY220 pKa = 7.69STKK223 pKa = 10.69AYY225 pKa = 8.43MSEE228 pKa = 3.62VMRR231 pKa = 11.84RR232 pKa = 11.84VGSYY236 pKa = 10.93DD237 pKa = 3.0RR238 pKa = 11.84TASGISSVFTSIAMMVQDD256 pKa = 5.48CINVVLRR263 pKa = 11.84RR264 pKa = 11.84FGVDD268 pKa = 3.71EE269 pKa = 4.0IQLIGEE275 pKa = 4.22AEE277 pKa = 4.1KK278 pKa = 10.65QVLEE282 pKa = 4.14WCRR285 pKa = 11.84QVDD288 pKa = 4.11DD289 pKa = 3.92VFKK292 pKa = 10.73IIDD295 pKa = 3.65TSTPQIKK302 pKa = 10.15DD303 pKa = 3.35LQFAQSLLAVGYY315 pKa = 9.09NFKK318 pKa = 10.75KK319 pKa = 9.53VTTASHH325 pKa = 7.13LLHH328 pKa = 6.68TIDD331 pKa = 4.33RR332 pKa = 11.84TLEE335 pKa = 4.02KK336 pKa = 10.71LNHH339 pKa = 6.55KK340 pKa = 9.9LVAHH344 pKa = 6.62KK345 pKa = 10.98GIMNDD350 pKa = 3.09EE351 pKa = 4.07NAFRR355 pKa = 11.84QQPILVMFGGGSGIGKK371 pKa = 8.32TNLIKK376 pKa = 10.84SLAGAILQLADD387 pKa = 3.28MCKK390 pKa = 9.9PEE392 pKa = 5.37HH393 pKa = 5.92VAQNLWQKK401 pKa = 11.43GDD403 pKa = 3.34TQYY406 pKa = 11.12WNGYY410 pKa = 8.05VGQLVYY416 pKa = 11.07IMDD419 pKa = 4.4DD420 pKa = 3.3VFQKK424 pKa = 11.04KK425 pKa = 7.96EE426 pKa = 3.85VAGGAEE432 pKa = 4.04NEE434 pKa = 4.15GFTIIRR440 pKa = 11.84AVSNWPFPLNFADD453 pKa = 3.99VEE455 pKa = 4.88SKK457 pKa = 10.88GRR459 pKa = 11.84FYY461 pKa = 10.92FSSKK465 pKa = 10.8LMIGTTNVDD474 pKa = 3.97DD475 pKa = 4.09IQRR478 pKa = 11.84AAGGVLQYY486 pKa = 10.63PEE488 pKa = 3.96AVVRR492 pKa = 11.84RR493 pKa = 11.84IEE495 pKa = 3.82HH496 pKa = 6.26GYY498 pKa = 9.05WLEE501 pKa = 4.27LADD504 pKa = 5.29DD505 pKa = 4.25YY506 pKa = 11.03KK507 pKa = 10.88TEE509 pKa = 4.45RR510 pKa = 11.84KK511 pKa = 7.5TLDD514 pKa = 3.13YY515 pKa = 11.35AKK517 pKa = 10.35LKK519 pKa = 10.19RR520 pKa = 11.84VHH522 pKa = 6.94DD523 pKa = 3.87EE524 pKa = 3.72RR525 pKa = 11.84LRR527 pKa = 11.84TLFADD532 pKa = 4.85GNQPTKK538 pKa = 10.47MEE540 pKa = 4.29VLRR543 pKa = 11.84SFPWEE548 pKa = 3.7AFEE551 pKa = 5.34ARR553 pKa = 11.84PHH555 pKa = 5.95NFASGAPGRR564 pKa = 11.84VGGFTSLLDD573 pKa = 3.35IAVEE577 pKa = 3.76IAEE580 pKa = 4.14EE581 pKa = 3.91LKK583 pKa = 10.91ARR585 pKa = 11.84SEE587 pKa = 4.02RR588 pKa = 11.84HH589 pKa = 4.6EE590 pKa = 4.57AEE592 pKa = 4.17CTMTLDD598 pKa = 2.96WLTAISRR605 pKa = 11.84ARR607 pKa = 11.84PEE609 pKa = 4.3AGFSSALIDD618 pKa = 3.5PTVGVSSQASHH629 pKa = 6.82SGEE632 pKa = 4.02PPQTGDD638 pKa = 3.57PPSSLRR644 pKa = 11.84GTMEE648 pKa = 3.69RR649 pKa = 11.84QIDD652 pKa = 3.86DD653 pKa = 3.64LVARR657 pKa = 11.84RR658 pKa = 11.84LRR660 pKa = 11.84ASTSTTPEE668 pKa = 3.22EE669 pKa = 4.07RR670 pKa = 11.84TAEE673 pKa = 4.07FAAGLDD679 pKa = 4.34GVAEE683 pKa = 4.07MCIGDD688 pKa = 3.98IKK690 pKa = 10.84RR691 pKa = 11.84EE692 pKa = 3.92QEE694 pKa = 3.72KK695 pKa = 10.44RR696 pKa = 11.84RR697 pKa = 11.84TAIEE701 pKa = 4.05YY702 pKa = 9.81IVYY705 pKa = 10.13LFKK708 pKa = 10.57TVAPSVLALLNVAMFLASGAISVWLHH734 pKa = 4.68YY735 pKa = 10.8KK736 pKa = 10.27FIQGICSLVSYY747 pKa = 10.5AFKK750 pKa = 10.78CVLSLMEE757 pKa = 4.12MARR760 pKa = 11.84DD761 pKa = 4.72FIFGTTPVEE770 pKa = 3.91QSVHH774 pKa = 5.69VEE776 pKa = 4.0HH777 pKa = 6.68KK778 pKa = 9.39TMPRR782 pKa = 11.84KK783 pKa = 8.72KK784 pKa = 8.24TQLAPPSAVIQMGNPPADD802 pKa = 3.49NLADD806 pKa = 3.22IAYY809 pKa = 8.55KK810 pKa = 10.53NSYY813 pKa = 10.47KK814 pKa = 10.53IMLGNGEE821 pKa = 4.08EE822 pKa = 4.17AKK824 pKa = 10.75SVGQVLMLRR833 pKa = 11.84QSFGVMPCHH842 pKa = 7.21FLTCPEE848 pKa = 4.24GPLRR852 pKa = 11.84FISCAGNGHH861 pKa = 6.43VIHH864 pKa = 7.09SSVQALKK871 pKa = 9.71ACRR874 pKa = 11.84VIALPTSDD882 pKa = 3.86MVFVDD887 pKa = 4.92LKK889 pKa = 10.33PILVRR894 pKa = 11.84AHH896 pKa = 6.64RR897 pKa = 11.84DD898 pKa = 3.18IVKK901 pKa = 10.34HH902 pKa = 5.53FVTDD906 pKa = 3.75KK907 pKa = 10.08TWNTLKK913 pKa = 10.86DD914 pKa = 3.42MRR916 pKa = 11.84NTSVRR921 pKa = 11.84LDD923 pKa = 3.52VARR926 pKa = 11.84MSGDD930 pKa = 3.26SLDD933 pKa = 3.5RR934 pKa = 11.84HH935 pKa = 5.88VMYY938 pKa = 10.83SSFLRR943 pKa = 11.84FDD945 pKa = 3.6PQITTIVTQVDD956 pKa = 3.84NVWAYY961 pKa = 10.43DD962 pKa = 3.79CPTKK966 pKa = 10.9VGDD969 pKa = 4.05CGAVLSISEE978 pKa = 3.94PCYY981 pKa = 10.58YY982 pKa = 10.27GGKK985 pKa = 10.04AILGIHH991 pKa = 6.54IAGKK995 pKa = 8.22TNSLMSVGSRR1005 pKa = 11.84EE1006 pKa = 4.17GYY1008 pKa = 9.93AAALTLEE1015 pKa = 5.02FIEE1018 pKa = 5.05KK1019 pKa = 10.54ALDD1022 pKa = 3.44KK1023 pKa = 10.76FKK1025 pKa = 9.57PTPVVDD1031 pKa = 4.38RR1032 pKa = 11.84FHH1034 pKa = 7.42EE1035 pKa = 4.51DD1036 pKa = 3.07LEE1038 pKa = 4.59GRR1040 pKa = 11.84GIEE1043 pKa = 4.08LEE1045 pKa = 4.06EE1046 pKa = 4.36VEE1048 pKa = 6.07DD1049 pKa = 5.3IEE1051 pKa = 4.68TQCGVVGGSITYY1063 pKa = 10.13VGSLSAAHH1071 pKa = 6.79SISQAGKK1078 pKa = 8.77SKK1080 pKa = 10.64LMRR1083 pKa = 11.84TGFSGWGRR1091 pKa = 11.84CPVAPAPLRR1100 pKa = 11.84PVYY1103 pKa = 10.38RR1104 pKa = 11.84DD1105 pKa = 3.72GILIKK1110 pKa = 10.53PMHH1113 pKa = 6.07QAMANYY1119 pKa = 8.05KK1120 pKa = 10.56KK1121 pKa = 9.88PVQPSAVRR1129 pKa = 11.84NGAAIMGLAMKK1140 pKa = 8.42MHH1142 pKa = 6.71NLHH1145 pKa = 6.39TKK1147 pKa = 8.97HH1148 pKa = 5.89CTRR1151 pKa = 11.84KK1152 pKa = 9.32ILTFEE1157 pKa = 4.14EE1158 pKa = 4.67SVLGVPEE1165 pKa = 4.06MKK1167 pKa = 10.16IKK1169 pKa = 9.18NTNRR1173 pKa = 11.84STSPGWPWRR1182 pKa = 11.84LEE1184 pKa = 4.11CANGKK1189 pKa = 9.64RR1190 pKa = 11.84DD1191 pKa = 3.38IFGSGVEE1198 pKa = 3.95FDD1200 pKa = 5.12LYY1202 pKa = 10.1TEE1204 pKa = 3.99KK1205 pKa = 11.18AMALRR1210 pKa = 11.84GRR1212 pKa = 11.84VEE1214 pKa = 4.27EE1215 pKa = 4.07VLEE1218 pKa = 3.95AAKK1221 pKa = 10.44RR1222 pKa = 11.84GEE1224 pKa = 4.06RR1225 pKa = 11.84LAHH1228 pKa = 5.76VFADD1232 pKa = 4.09FLKK1235 pKa = 11.09DD1236 pKa = 3.15EE1237 pKa = 4.61TRR1239 pKa = 11.84PHH1241 pKa = 6.36AKK1243 pKa = 9.83VEE1245 pKa = 4.15QVMTRR1250 pKa = 11.84AISGAPLDD1258 pKa = 3.68YY1259 pKa = 10.52TIAVRR1264 pKa = 11.84MYY1266 pKa = 9.38FGAFLSSMFLHH1277 pKa = 5.79NTVSGMAPGLNYY1289 pKa = 10.64YY1290 pKa = 10.51SDD1292 pKa = 3.41WSTLARR1298 pKa = 11.84EE1299 pKa = 4.47LLRR1302 pKa = 11.84KK1303 pKa = 8.78GEE1305 pKa = 4.11KK1306 pKa = 9.41MFAGDD1311 pKa = 4.74FKK1313 pKa = 11.52AFDD1316 pKa = 3.88ASEE1319 pKa = 4.07QPDD1322 pKa = 3.4IHH1324 pKa = 8.36LLILDD1329 pKa = 5.11YY1330 pKa = 11.36INDD1333 pKa = 3.56WYY1335 pKa = 10.47DD1336 pKa = 3.15QYY1338 pKa = 11.92GADD1341 pKa = 3.53PEE1343 pKa = 4.39GRR1345 pKa = 11.84RR1346 pKa = 11.84VRR1348 pKa = 11.84TVLFEE1353 pKa = 4.45DD1354 pKa = 5.61LIHH1357 pKa = 6.46SRR1359 pKa = 11.84HH1360 pKa = 5.39LTGDD1364 pKa = 3.51SYY1366 pKa = 12.01KK1367 pKa = 10.68LDD1369 pKa = 5.11TIVQWNKK1376 pKa = 10.34SLPSGHH1382 pKa = 7.46PLTTAVNSMYY1392 pKa = 11.18SLFTLTACYY1401 pKa = 10.47VHH1403 pKa = 6.42LTKK1406 pKa = 10.81DD1407 pKa = 3.55HH1408 pKa = 6.38EE1409 pKa = 4.53NMWDD1413 pKa = 3.37HH1414 pKa = 7.04VFICTFGDD1422 pKa = 3.91DD1423 pKa = 3.99NVVSADD1429 pKa = 3.99DD1430 pKa = 3.5DD1431 pKa = 4.41TIAVFNQVSVAKK1443 pKa = 10.59AMQEE1447 pKa = 4.17LFHH1450 pKa = 6.4LTYY1453 pKa = 10.73TSDD1456 pKa = 3.42KK1457 pKa = 10.59KK1458 pKa = 10.89DD1459 pKa = 3.47AEE1461 pKa = 4.28LVPYY1465 pKa = 7.85EE1466 pKa = 4.53TIHH1469 pKa = 7.82DD1470 pKa = 3.58ITFLKK1475 pKa = 10.48RR1476 pKa = 11.84SFVRR1480 pKa = 11.84SQSDD1484 pKa = 3.68GGWIAPLAMDD1494 pKa = 4.74SILYY1498 pKa = 8.06RR1499 pKa = 11.84TYY1501 pKa = 10.63FYY1503 pKa = 10.95QNPRR1507 pKa = 11.84NYY1509 pKa = 9.97VRR1511 pKa = 11.84DD1512 pKa = 3.1QATNFKK1518 pKa = 9.18EE1519 pKa = 4.05ALMEE1523 pKa = 4.3LSLHH1527 pKa = 6.87GEE1529 pKa = 4.31EE1530 pKa = 4.69EE1531 pKa = 3.75WDD1533 pKa = 3.74EE1534 pKa = 4.3RR1535 pKa = 11.84FQAAAVYY1542 pKa = 10.54CRR1544 pKa = 11.84DD1545 pKa = 3.56NNIDD1549 pKa = 3.41FCITSRR1555 pKa = 11.84EE1556 pKa = 4.24QARR1559 pKa = 11.84QMCLARR1565 pKa = 11.84TDD1567 pKa = 3.03VWFF1570 pKa = 4.46
MM1 pKa = 7.62SFTTNEE7 pKa = 3.99SEE9 pKa = 4.36VVVNVKK15 pKa = 9.99LLRR18 pKa = 11.84RR19 pKa = 11.84AEE21 pKa = 3.91RR22 pKa = 11.84EE23 pKa = 4.09AIQRR27 pKa = 11.84ACNIRR32 pKa = 11.84RR33 pKa = 11.84LKK35 pKa = 10.64KK36 pKa = 10.4AFKK39 pKa = 10.38SLPKK43 pKa = 10.1YY44 pKa = 9.98EE45 pKa = 4.18RR46 pKa = 11.84EE47 pKa = 3.89QVIASARR54 pKa = 11.84DD55 pKa = 3.27AKK57 pKa = 10.95YY58 pKa = 10.34SFVRR62 pKa = 11.84EE63 pKa = 3.82EE64 pKa = 4.24SGITDD69 pKa = 3.85YY70 pKa = 11.4LYY72 pKa = 11.31GAAAVAAGAAGAAAGFSIAKK92 pKa = 8.23TSHH95 pKa = 6.63EE96 pKa = 3.89IGEE99 pKa = 4.37AVKK102 pKa = 9.7DD103 pKa = 3.71TLSGVNAVMQPAGAIANSVGGALEE127 pKa = 3.93YY128 pKa = 10.56LYY130 pKa = 11.34SLVKK134 pKa = 10.11QVYY137 pKa = 7.49DD138 pKa = 3.42TCANIVGKK146 pKa = 10.05FSQLIFGIFVVAMKK160 pKa = 10.7ACFGLVNMASRR171 pKa = 11.84LFDD174 pKa = 3.53SVVSMVAPDD183 pKa = 3.58FHH185 pKa = 8.52LLDD188 pKa = 4.14SLEE191 pKa = 4.4EE192 pKa = 4.87DD193 pKa = 4.12DD194 pKa = 5.93VLPQSGCMSYY204 pKa = 10.8LPQVVTLVCTFFVPTYY220 pKa = 7.69STKK223 pKa = 10.69AYY225 pKa = 8.43MSEE228 pKa = 3.62VMRR231 pKa = 11.84RR232 pKa = 11.84VGSYY236 pKa = 10.93DD237 pKa = 3.0RR238 pKa = 11.84TASGISSVFTSIAMMVQDD256 pKa = 5.48CINVVLRR263 pKa = 11.84RR264 pKa = 11.84FGVDD268 pKa = 3.71EE269 pKa = 4.0IQLIGEE275 pKa = 4.22AEE277 pKa = 4.1KK278 pKa = 10.65QVLEE282 pKa = 4.14WCRR285 pKa = 11.84QVDD288 pKa = 4.11DD289 pKa = 3.92VFKK292 pKa = 10.73IIDD295 pKa = 3.65TSTPQIKK302 pKa = 10.15DD303 pKa = 3.35LQFAQSLLAVGYY315 pKa = 9.09NFKK318 pKa = 10.75KK319 pKa = 9.53VTTASHH325 pKa = 7.13LLHH328 pKa = 6.68TIDD331 pKa = 4.33RR332 pKa = 11.84TLEE335 pKa = 4.02KK336 pKa = 10.71LNHH339 pKa = 6.55KK340 pKa = 9.9LVAHH344 pKa = 6.62KK345 pKa = 10.98GIMNDD350 pKa = 3.09EE351 pKa = 4.07NAFRR355 pKa = 11.84QQPILVMFGGGSGIGKK371 pKa = 8.32TNLIKK376 pKa = 10.84SLAGAILQLADD387 pKa = 3.28MCKK390 pKa = 9.9PEE392 pKa = 5.37HH393 pKa = 5.92VAQNLWQKK401 pKa = 11.43GDD403 pKa = 3.34TQYY406 pKa = 11.12WNGYY410 pKa = 8.05VGQLVYY416 pKa = 11.07IMDD419 pKa = 4.4DD420 pKa = 3.3VFQKK424 pKa = 11.04KK425 pKa = 7.96EE426 pKa = 3.85VAGGAEE432 pKa = 4.04NEE434 pKa = 4.15GFTIIRR440 pKa = 11.84AVSNWPFPLNFADD453 pKa = 3.99VEE455 pKa = 4.88SKK457 pKa = 10.88GRR459 pKa = 11.84FYY461 pKa = 10.92FSSKK465 pKa = 10.8LMIGTTNVDD474 pKa = 3.97DD475 pKa = 4.09IQRR478 pKa = 11.84AAGGVLQYY486 pKa = 10.63PEE488 pKa = 3.96AVVRR492 pKa = 11.84RR493 pKa = 11.84IEE495 pKa = 3.82HH496 pKa = 6.26GYY498 pKa = 9.05WLEE501 pKa = 4.27LADD504 pKa = 5.29DD505 pKa = 4.25YY506 pKa = 11.03KK507 pKa = 10.88TEE509 pKa = 4.45RR510 pKa = 11.84KK511 pKa = 7.5TLDD514 pKa = 3.13YY515 pKa = 11.35AKK517 pKa = 10.35LKK519 pKa = 10.19RR520 pKa = 11.84VHH522 pKa = 6.94DD523 pKa = 3.87EE524 pKa = 3.72RR525 pKa = 11.84LRR527 pKa = 11.84TLFADD532 pKa = 4.85GNQPTKK538 pKa = 10.47MEE540 pKa = 4.29VLRR543 pKa = 11.84SFPWEE548 pKa = 3.7AFEE551 pKa = 5.34ARR553 pKa = 11.84PHH555 pKa = 5.95NFASGAPGRR564 pKa = 11.84VGGFTSLLDD573 pKa = 3.35IAVEE577 pKa = 3.76IAEE580 pKa = 4.14EE581 pKa = 3.91LKK583 pKa = 10.91ARR585 pKa = 11.84SEE587 pKa = 4.02RR588 pKa = 11.84HH589 pKa = 4.6EE590 pKa = 4.57AEE592 pKa = 4.17CTMTLDD598 pKa = 2.96WLTAISRR605 pKa = 11.84ARR607 pKa = 11.84PEE609 pKa = 4.3AGFSSALIDD618 pKa = 3.5PTVGVSSQASHH629 pKa = 6.82SGEE632 pKa = 4.02PPQTGDD638 pKa = 3.57PPSSLRR644 pKa = 11.84GTMEE648 pKa = 3.69RR649 pKa = 11.84QIDD652 pKa = 3.86DD653 pKa = 3.64LVARR657 pKa = 11.84RR658 pKa = 11.84LRR660 pKa = 11.84ASTSTTPEE668 pKa = 3.22EE669 pKa = 4.07RR670 pKa = 11.84TAEE673 pKa = 4.07FAAGLDD679 pKa = 4.34GVAEE683 pKa = 4.07MCIGDD688 pKa = 3.98IKK690 pKa = 10.84RR691 pKa = 11.84EE692 pKa = 3.92QEE694 pKa = 3.72KK695 pKa = 10.44RR696 pKa = 11.84RR697 pKa = 11.84TAIEE701 pKa = 4.05YY702 pKa = 9.81IVYY705 pKa = 10.13LFKK708 pKa = 10.57TVAPSVLALLNVAMFLASGAISVWLHH734 pKa = 4.68YY735 pKa = 10.8KK736 pKa = 10.27FIQGICSLVSYY747 pKa = 10.5AFKK750 pKa = 10.78CVLSLMEE757 pKa = 4.12MARR760 pKa = 11.84DD761 pKa = 4.72FIFGTTPVEE770 pKa = 3.91QSVHH774 pKa = 5.69VEE776 pKa = 4.0HH777 pKa = 6.68KK778 pKa = 9.39TMPRR782 pKa = 11.84KK783 pKa = 8.72KK784 pKa = 8.24TQLAPPSAVIQMGNPPADD802 pKa = 3.49NLADD806 pKa = 3.22IAYY809 pKa = 8.55KK810 pKa = 10.53NSYY813 pKa = 10.47KK814 pKa = 10.53IMLGNGEE821 pKa = 4.08EE822 pKa = 4.17AKK824 pKa = 10.75SVGQVLMLRR833 pKa = 11.84QSFGVMPCHH842 pKa = 7.21FLTCPEE848 pKa = 4.24GPLRR852 pKa = 11.84FISCAGNGHH861 pKa = 6.43VIHH864 pKa = 7.09SSVQALKK871 pKa = 9.71ACRR874 pKa = 11.84VIALPTSDD882 pKa = 3.86MVFVDD887 pKa = 4.92LKK889 pKa = 10.33PILVRR894 pKa = 11.84AHH896 pKa = 6.64RR897 pKa = 11.84DD898 pKa = 3.18IVKK901 pKa = 10.34HH902 pKa = 5.53FVTDD906 pKa = 3.75KK907 pKa = 10.08TWNTLKK913 pKa = 10.86DD914 pKa = 3.42MRR916 pKa = 11.84NTSVRR921 pKa = 11.84LDD923 pKa = 3.52VARR926 pKa = 11.84MSGDD930 pKa = 3.26SLDD933 pKa = 3.5RR934 pKa = 11.84HH935 pKa = 5.88VMYY938 pKa = 10.83SSFLRR943 pKa = 11.84FDD945 pKa = 3.6PQITTIVTQVDD956 pKa = 3.84NVWAYY961 pKa = 10.43DD962 pKa = 3.79CPTKK966 pKa = 10.9VGDD969 pKa = 4.05CGAVLSISEE978 pKa = 3.94PCYY981 pKa = 10.58YY982 pKa = 10.27GGKK985 pKa = 10.04AILGIHH991 pKa = 6.54IAGKK995 pKa = 8.22TNSLMSVGSRR1005 pKa = 11.84EE1006 pKa = 4.17GYY1008 pKa = 9.93AAALTLEE1015 pKa = 5.02FIEE1018 pKa = 5.05KK1019 pKa = 10.54ALDD1022 pKa = 3.44KK1023 pKa = 10.76FKK1025 pKa = 9.57PTPVVDD1031 pKa = 4.38RR1032 pKa = 11.84FHH1034 pKa = 7.42EE1035 pKa = 4.51DD1036 pKa = 3.07LEE1038 pKa = 4.59GRR1040 pKa = 11.84GIEE1043 pKa = 4.08LEE1045 pKa = 4.06EE1046 pKa = 4.36VEE1048 pKa = 6.07DD1049 pKa = 5.3IEE1051 pKa = 4.68TQCGVVGGSITYY1063 pKa = 10.13VGSLSAAHH1071 pKa = 6.79SISQAGKK1078 pKa = 8.77SKK1080 pKa = 10.64LMRR1083 pKa = 11.84TGFSGWGRR1091 pKa = 11.84CPVAPAPLRR1100 pKa = 11.84PVYY1103 pKa = 10.38RR1104 pKa = 11.84DD1105 pKa = 3.72GILIKK1110 pKa = 10.53PMHH1113 pKa = 6.07QAMANYY1119 pKa = 8.05KK1120 pKa = 10.56KK1121 pKa = 9.88PVQPSAVRR1129 pKa = 11.84NGAAIMGLAMKK1140 pKa = 8.42MHH1142 pKa = 6.71NLHH1145 pKa = 6.39TKK1147 pKa = 8.97HH1148 pKa = 5.89CTRR1151 pKa = 11.84KK1152 pKa = 9.32ILTFEE1157 pKa = 4.14EE1158 pKa = 4.67SVLGVPEE1165 pKa = 4.06MKK1167 pKa = 10.16IKK1169 pKa = 9.18NTNRR1173 pKa = 11.84STSPGWPWRR1182 pKa = 11.84LEE1184 pKa = 4.11CANGKK1189 pKa = 9.64RR1190 pKa = 11.84DD1191 pKa = 3.38IFGSGVEE1198 pKa = 3.95FDD1200 pKa = 5.12LYY1202 pKa = 10.1TEE1204 pKa = 3.99KK1205 pKa = 11.18AMALRR1210 pKa = 11.84GRR1212 pKa = 11.84VEE1214 pKa = 4.27EE1215 pKa = 4.07VLEE1218 pKa = 3.95AAKK1221 pKa = 10.44RR1222 pKa = 11.84GEE1224 pKa = 4.06RR1225 pKa = 11.84LAHH1228 pKa = 5.76VFADD1232 pKa = 4.09FLKK1235 pKa = 11.09DD1236 pKa = 3.15EE1237 pKa = 4.61TRR1239 pKa = 11.84PHH1241 pKa = 6.36AKK1243 pKa = 9.83VEE1245 pKa = 4.15QVMTRR1250 pKa = 11.84AISGAPLDD1258 pKa = 3.68YY1259 pKa = 10.52TIAVRR1264 pKa = 11.84MYY1266 pKa = 9.38FGAFLSSMFLHH1277 pKa = 5.79NTVSGMAPGLNYY1289 pKa = 10.64YY1290 pKa = 10.51SDD1292 pKa = 3.41WSTLARR1298 pKa = 11.84EE1299 pKa = 4.47LLRR1302 pKa = 11.84KK1303 pKa = 8.78GEE1305 pKa = 4.11KK1306 pKa = 9.41MFAGDD1311 pKa = 4.74FKK1313 pKa = 11.52AFDD1316 pKa = 3.88ASEE1319 pKa = 4.07QPDD1322 pKa = 3.4IHH1324 pKa = 8.36LLILDD1329 pKa = 5.11YY1330 pKa = 11.36INDD1333 pKa = 3.56WYY1335 pKa = 10.47DD1336 pKa = 3.15QYY1338 pKa = 11.92GADD1341 pKa = 3.53PEE1343 pKa = 4.39GRR1345 pKa = 11.84RR1346 pKa = 11.84VRR1348 pKa = 11.84TVLFEE1353 pKa = 4.45DD1354 pKa = 5.61LIHH1357 pKa = 6.46SRR1359 pKa = 11.84HH1360 pKa = 5.39LTGDD1364 pKa = 3.51SYY1366 pKa = 12.01KK1367 pKa = 10.68LDD1369 pKa = 5.11TIVQWNKK1376 pKa = 10.34SLPSGHH1382 pKa = 7.46PLTTAVNSMYY1392 pKa = 11.18SLFTLTACYY1401 pKa = 10.47VHH1403 pKa = 6.42LTKK1406 pKa = 10.81DD1407 pKa = 3.55HH1408 pKa = 6.38EE1409 pKa = 4.53NMWDD1413 pKa = 3.37HH1414 pKa = 7.04VFICTFGDD1422 pKa = 3.91DD1423 pKa = 3.99NVVSADD1429 pKa = 3.99DD1430 pKa = 3.5DD1431 pKa = 4.41TIAVFNQVSVAKK1443 pKa = 10.59AMQEE1447 pKa = 4.17LFHH1450 pKa = 6.4LTYY1453 pKa = 10.73TSDD1456 pKa = 3.42KK1457 pKa = 10.59KK1458 pKa = 10.89DD1459 pKa = 3.47AEE1461 pKa = 4.28LVPYY1465 pKa = 7.85EE1466 pKa = 4.53TIHH1469 pKa = 7.82DD1470 pKa = 3.58ITFLKK1475 pKa = 10.48RR1476 pKa = 11.84SFVRR1480 pKa = 11.84SQSDD1484 pKa = 3.68GGWIAPLAMDD1494 pKa = 4.74SILYY1498 pKa = 8.06RR1499 pKa = 11.84TYY1501 pKa = 10.63FYY1503 pKa = 10.95QNPRR1507 pKa = 11.84NYY1509 pKa = 9.97VRR1511 pKa = 11.84DD1512 pKa = 3.1QATNFKK1518 pKa = 9.18EE1519 pKa = 4.05ALMEE1523 pKa = 4.3LSLHH1527 pKa = 6.87GEE1529 pKa = 4.31EE1530 pKa = 4.69EE1531 pKa = 3.75WDD1533 pKa = 3.74EE1534 pKa = 4.3RR1535 pKa = 11.84FQAAAVYY1542 pKa = 10.54CRR1544 pKa = 11.84DD1545 pKa = 3.56NNIDD1549 pKa = 3.41FCITSRR1555 pKa = 11.84EE1556 pKa = 4.24QARR1559 pKa = 11.84QMCLARR1565 pKa = 11.84TDD1567 pKa = 3.03VWFF1570 pKa = 4.46
Molecular weight: 174.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMD5|A0A1L3KMD5_9VIRU Uncharacterized protein OS=Wenzhou picorna-like virus 33 OX=1923619 PE=4 SV=1
MM1 pKa = 7.59LSGVLATASRR11 pKa = 11.84VPKK14 pKa = 10.42AVGRR18 pKa = 11.84LFPSLAPIMGSTSWFLAASARR39 pKa = 11.84AANAFGFSKK48 pKa = 10.52PVVQKK53 pKa = 10.26VPPVLRR59 pKa = 11.84TFHH62 pKa = 6.76NYY64 pKa = 8.98EE65 pKa = 4.3SNIDD69 pKa = 3.64MPVPAVAVGAFAGNSVAVSKK89 pKa = 11.11ALGGTDD95 pKa = 3.18IDD97 pKa = 4.44EE98 pKa = 4.31MSFDD102 pKa = 4.59YY103 pKa = 10.76ILSKK107 pKa = 10.35PSQIFRR113 pKa = 11.84GTMSTTDD120 pKa = 2.79AHH122 pKa = 6.0ATVEE126 pKa = 4.47YY127 pKa = 9.86ASHH130 pKa = 6.3VALTHH135 pKa = 4.44MWFRR139 pKa = 11.84APTTGVDD146 pKa = 3.06RR147 pKa = 11.84GNISFPRR154 pKa = 11.84GAPAVGGTTAVIPSNLLYY172 pKa = 10.22FAQHH176 pKa = 5.09FRR178 pKa = 11.84LWHH181 pKa = 6.1GDD183 pKa = 2.95LVYY186 pKa = 10.58RR187 pKa = 11.84VTFAKK192 pKa = 10.43SKK194 pKa = 8.65FHH196 pKa = 6.64TGRR199 pKa = 11.84VMFSFIPNYY208 pKa = 10.04QRR210 pKa = 11.84VTNVSTYY217 pKa = 10.95SEE219 pKa = 4.46AGLAGGPAPGVFAGDD234 pKa = 4.27LQPSQYY240 pKa = 11.5SLVFDD245 pKa = 5.06LKK247 pKa = 11.0DD248 pKa = 3.37GSEE251 pKa = 4.56FEE253 pKa = 4.92FEE255 pKa = 4.39VPYY258 pKa = 9.94IAPMPHH264 pKa = 6.95VGINDD269 pKa = 3.96SIGFVTMQIMDD280 pKa = 4.4PLVANGEE287 pKa = 4.24SSDD290 pKa = 3.78TISFIVEE297 pKa = 3.66VAAKK301 pKa = 9.82PGFYY305 pKa = 10.1FAGIAGPNVPIWPDD319 pKa = 3.2LTAPTIEE326 pKa = 4.29FQSGVGGITVDD337 pKa = 3.82ASQHH341 pKa = 5.06SVGEE345 pKa = 4.21KK346 pKa = 10.4FMSAKK351 pKa = 10.11QLMLHH356 pKa = 6.84PIMRR360 pKa = 11.84RR361 pKa = 11.84FSHH364 pKa = 6.77ANNTTAQGTIPLWPCSPAWGDD385 pKa = 3.74GAPLPVNTQRR395 pKa = 11.84QYY397 pKa = 10.41PFIRR401 pKa = 11.84SGMVAQCFAFGIGSTLLHH419 pKa = 6.2MQTAGLSVSSIVRR432 pKa = 11.84VVQSPGDD439 pKa = 3.44NGEE442 pKa = 4.24AVTGTVPGLYY452 pKa = 9.44NRR454 pKa = 11.84PIASPNTAWAQSSRR468 pKa = 11.84GPTSTEE474 pKa = 3.92SYY476 pKa = 10.1LLPMLSAAPRR486 pKa = 11.84FRR488 pKa = 11.84IGDD491 pKa = 3.79FNNASPTRR499 pKa = 11.84DD500 pKa = 3.16WSPGGSSTISTDD512 pKa = 2.98SLAVKK517 pKa = 7.42TTYY520 pKa = 9.89NWSVANVDD528 pKa = 3.07GATRR532 pKa = 11.84LWSWGVSAADD542 pKa = 4.62DD543 pKa = 3.92ARR545 pKa = 11.84CAAWIGPCPMVLANNGSTTNTFYY568 pKa = 10.67IDD570 pKa = 3.38GGVNNN575 pKa = 4.77
MM1 pKa = 7.59LSGVLATASRR11 pKa = 11.84VPKK14 pKa = 10.42AVGRR18 pKa = 11.84LFPSLAPIMGSTSWFLAASARR39 pKa = 11.84AANAFGFSKK48 pKa = 10.52PVVQKK53 pKa = 10.26VPPVLRR59 pKa = 11.84TFHH62 pKa = 6.76NYY64 pKa = 8.98EE65 pKa = 4.3SNIDD69 pKa = 3.64MPVPAVAVGAFAGNSVAVSKK89 pKa = 11.11ALGGTDD95 pKa = 3.18IDD97 pKa = 4.44EE98 pKa = 4.31MSFDD102 pKa = 4.59YY103 pKa = 10.76ILSKK107 pKa = 10.35PSQIFRR113 pKa = 11.84GTMSTTDD120 pKa = 2.79AHH122 pKa = 6.0ATVEE126 pKa = 4.47YY127 pKa = 9.86ASHH130 pKa = 6.3VALTHH135 pKa = 4.44MWFRR139 pKa = 11.84APTTGVDD146 pKa = 3.06RR147 pKa = 11.84GNISFPRR154 pKa = 11.84GAPAVGGTTAVIPSNLLYY172 pKa = 10.22FAQHH176 pKa = 5.09FRR178 pKa = 11.84LWHH181 pKa = 6.1GDD183 pKa = 2.95LVYY186 pKa = 10.58RR187 pKa = 11.84VTFAKK192 pKa = 10.43SKK194 pKa = 8.65FHH196 pKa = 6.64TGRR199 pKa = 11.84VMFSFIPNYY208 pKa = 10.04QRR210 pKa = 11.84VTNVSTYY217 pKa = 10.95SEE219 pKa = 4.46AGLAGGPAPGVFAGDD234 pKa = 4.27LQPSQYY240 pKa = 11.5SLVFDD245 pKa = 5.06LKK247 pKa = 11.0DD248 pKa = 3.37GSEE251 pKa = 4.56FEE253 pKa = 4.92FEE255 pKa = 4.39VPYY258 pKa = 9.94IAPMPHH264 pKa = 6.95VGINDD269 pKa = 3.96SIGFVTMQIMDD280 pKa = 4.4PLVANGEE287 pKa = 4.24SSDD290 pKa = 3.78TISFIVEE297 pKa = 3.66VAAKK301 pKa = 9.82PGFYY305 pKa = 10.1FAGIAGPNVPIWPDD319 pKa = 3.2LTAPTIEE326 pKa = 4.29FQSGVGGITVDD337 pKa = 3.82ASQHH341 pKa = 5.06SVGEE345 pKa = 4.21KK346 pKa = 10.4FMSAKK351 pKa = 10.11QLMLHH356 pKa = 6.84PIMRR360 pKa = 11.84RR361 pKa = 11.84FSHH364 pKa = 6.77ANNTTAQGTIPLWPCSPAWGDD385 pKa = 3.74GAPLPVNTQRR395 pKa = 11.84QYY397 pKa = 10.41PFIRR401 pKa = 11.84SGMVAQCFAFGIGSTLLHH419 pKa = 6.2MQTAGLSVSSIVRR432 pKa = 11.84VVQSPGDD439 pKa = 3.44NGEE442 pKa = 4.24AVTGTVPGLYY452 pKa = 9.44NRR454 pKa = 11.84PIASPNTAWAQSSRR468 pKa = 11.84GPTSTEE474 pKa = 3.92SYY476 pKa = 10.1LLPMLSAAPRR486 pKa = 11.84FRR488 pKa = 11.84IGDD491 pKa = 3.79FNNASPTRR499 pKa = 11.84DD500 pKa = 3.16WSPGGSSTISTDD512 pKa = 2.98SLAVKK517 pKa = 7.42TTYY520 pKa = 9.89NWSVANVDD528 pKa = 3.07GATRR532 pKa = 11.84LWSWGVSAADD542 pKa = 4.62DD543 pKa = 3.92ARR545 pKa = 11.84CAAWIGPCPMVLANNGSTTNTFYY568 pKa = 10.67IDD570 pKa = 3.38GGVNNN575 pKa = 4.77
Molecular weight: 61.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2145 |
575 |
1570 |
1072.5 |
118.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.604 ± 0.634 | 1.492 ± 0.428 |
5.361 ± 0.638 | 4.988 ± 1.466 |
4.942 ± 0.429 | 7.319 ± 1.02 |
2.471 ± 0.206 | 5.175 ± 0.164 |
4.382 ± 1.234 | 7.646 ± 1.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.17 ± 0.115 | 3.543 ± 0.526 |
4.848 ± 1.507 | 3.31 ± 0.19 |
5.315 ± 0.613 | 7.646 ± 1.126 |
6.107 ± 0.644 | 8.065 ± 0.152 |
1.492 ± 0.32 | 3.124 ± 0.277 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
