
Acanthochromis polyacanthus (spiny chromis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai;
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
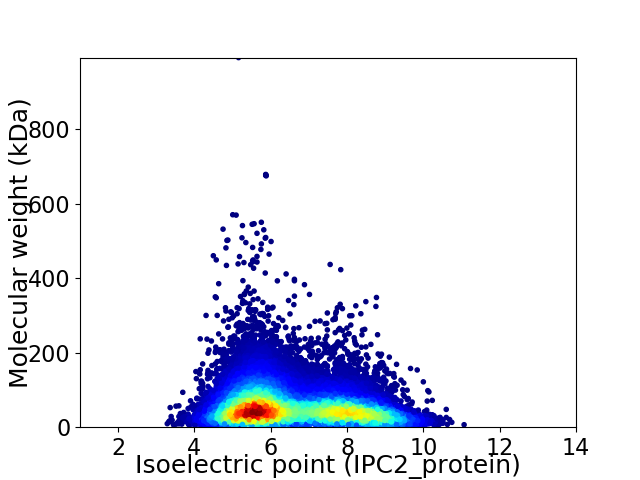
Virtual 2D-PAGE plot for 33480 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q1H447|A0A3Q1H447_9TELE Zinc finger protein 556-like OS=Acanthochromis polyacanthus OX=80966 PE=4 SV=1
TT1 pKa = 7.16IAEE4 pKa = 4.77CLPCGKK10 pKa = 7.74QHH12 pKa = 7.23KK13 pKa = 10.58NFDD16 pKa = 4.47LDD18 pKa = 3.66PAPLIKK24 pKa = 10.42CPPGTCFRR32 pKa = 11.84QNMCIPSNSGRR43 pKa = 11.84FQCAPCPDD51 pKa = 4.17GFTGDD56 pKa = 4.38GVHH59 pKa = 7.02CDD61 pKa = 5.04DD62 pKa = 5.32VDD64 pKa = 3.52EE65 pKa = 5.54CKK67 pKa = 10.62FNPCFPGVRR76 pKa = 11.84CVNTAPGFRR85 pKa = 11.84CEE87 pKa = 4.09KK88 pKa = 10.56CPLGYY93 pKa = 9.12TGPLLTGVGVSYY105 pKa = 11.38AKK107 pKa = 9.9TNKK110 pKa = 9.05QVCEE114 pKa = 5.17DD115 pKa = 3.46IDD117 pKa = 3.98EE118 pKa = 4.7CLGPPDD124 pKa = 5.25NGGCTANSDD133 pKa = 3.94CHH135 pKa = 5.11NTMGSFRR142 pKa = 11.84CGNCKK147 pKa = 10.33SGFTGDD153 pKa = 3.73QVRR156 pKa = 11.84GCHH159 pKa = 6.9GDD161 pKa = 3.47RR162 pKa = 11.84LCPNGQPNPCDD173 pKa = 3.35INAEE177 pKa = 4.41CIVEE181 pKa = 4.0RR182 pKa = 11.84DD183 pKa = 3.56GSISCACGVGWAGNGYY199 pKa = 9.96VCGKK203 pKa = 9.08DD204 pKa = 3.26TDD206 pKa = 4.13IDD208 pKa = 3.69AHH210 pKa = 6.92PDD212 pKa = 3.21DD213 pKa = 4.71TLQCRR218 pKa = 11.84DD219 pKa = 3.77STCKK223 pKa = 9.81QDD225 pKa = 3.25NCVFVPNSGQEE236 pKa = 3.9DD237 pKa = 4.01ADD239 pKa = 4.23RR240 pKa = 11.84DD241 pKa = 4.12GMGDD245 pKa = 3.44ACDD248 pKa = 5.25DD249 pKa = 4.49DD250 pKa = 6.68ADD252 pKa = 4.25SDD254 pKa = 4.63GIMNIDD260 pKa = 3.79DD261 pKa = 4.27NCWLVPNVNQKK272 pKa = 10.44NSDD275 pKa = 3.57KK276 pKa = 10.78DD277 pKa = 3.66LHH279 pKa = 7.57GDD281 pKa = 3.17ACDD284 pKa = 3.37NCKK287 pKa = 9.07TVEE290 pKa = 4.15NPSQRR295 pKa = 11.84DD296 pKa = 3.21TDD298 pKa = 3.54QDD300 pKa = 3.95GLGDD304 pKa = 3.9EE305 pKa = 5.58CDD307 pKa = 4.71DD308 pKa = 5.76DD309 pKa = 4.25MDD311 pKa = 6.43GDD313 pKa = 4.13GLKK316 pKa = 10.81NILDD320 pKa = 3.68NCQRR324 pKa = 11.84VPNLDD329 pKa = 3.29QKK331 pKa = 11.54DD332 pKa = 3.66RR333 pKa = 11.84DD334 pKa = 3.67NDD336 pKa = 4.11GVGDD340 pKa = 4.22ACDD343 pKa = 3.93SCPDD347 pKa = 3.54MANPNQSDD355 pKa = 3.51SDD357 pKa = 4.12DD358 pKa = 4.47DD359 pKa = 4.96LVGDD363 pKa = 4.17TCDD366 pKa = 5.48DD367 pKa = 4.73NIDD370 pKa = 3.69SDD372 pKa = 4.95GDD374 pKa = 3.6GHH376 pKa = 7.04QNTKK380 pKa = 10.88DD381 pKa = 3.38NCPDD385 pKa = 3.72VINSSQLDD393 pKa = 3.62TDD395 pKa = 3.7KK396 pKa = 11.69DD397 pKa = 4.0GMGDD401 pKa = 3.49EE402 pKa = 5.68CDD404 pKa = 5.62DD405 pKa = 5.68DD406 pKa = 6.32DD407 pKa = 7.09DD408 pKa = 6.04NDD410 pKa = 5.26GILDD414 pKa = 3.81EE415 pKa = 6.04DD416 pKa = 4.7DD417 pKa = 3.7NCRR420 pKa = 11.84LVPNPDD426 pKa = 3.22QEE428 pKa = 4.67DD429 pKa = 3.77ANKK432 pKa = 10.9DD433 pKa = 3.63KK434 pKa = 11.66VGDD437 pKa = 3.7ACEE440 pKa = 4.28GDD442 pKa = 3.48FDD444 pKa = 4.85KK445 pKa = 11.65DD446 pKa = 3.6NVIDD450 pKa = 5.66IIDD453 pKa = 4.07HH454 pKa = 6.03CPEE457 pKa = 3.9NAEE460 pKa = 4.03VTLTDD465 pKa = 3.81FRR467 pKa = 11.84AYY469 pKa = 8.19QTVVLDD475 pKa = 4.24PEE477 pKa = 5.01GDD479 pKa = 3.9SQIDD483 pKa = 3.89PNWVVLNQGMEE494 pKa = 4.11IVQTMNSDD502 pKa = 3.19PGLAVGYY509 pKa = 9.83KK510 pKa = 10.02AFSGVDD516 pKa = 3.74FEE518 pKa = 5.24GTFHH522 pKa = 6.81VNTVTDD528 pKa = 3.57DD529 pKa = 4.08DD530 pKa = 4.35YY531 pKa = 12.0AGFIFGYY538 pKa = 9.58QDD540 pKa = 2.83SSSFYY545 pKa = 10.47VVMWKK550 pKa = 7.78QTEE553 pKa = 3.84QTYY556 pKa = 8.18WQAAPFRR563 pKa = 11.84AVADD567 pKa = 3.86PGIQLKK573 pKa = 10.07AVKK576 pKa = 10.09SKK578 pKa = 9.5TGPGEE583 pKa = 3.83YY584 pKa = 10.03LRR586 pKa = 11.84NSLWHH591 pKa = 6.45TGNTPDD597 pKa = 5.06QVRR600 pKa = 11.84LLWKK604 pKa = 10.3DD605 pKa = 3.22PRR607 pKa = 11.84NVGWKK612 pKa = 10.39DD613 pKa = 3.18KK614 pKa = 11.21VSYY617 pKa = 10.33RR618 pKa = 11.84WFLQHH623 pKa = 6.81RR624 pKa = 11.84PQVGYY629 pKa = 9.82IRR631 pKa = 11.84ARR633 pKa = 11.84FFEE636 pKa = 4.37GSNLVADD643 pKa = 4.25TGMITDD649 pKa = 3.77TSMRR653 pKa = 11.84GGRR656 pKa = 11.84LGVFCFSQEE665 pKa = 3.94NIIWSNLKK673 pKa = 9.68YY674 pKa = 10.31RR675 pKa = 11.84CNDD678 pKa = 4.36TIPADD683 pKa = 3.95YY684 pKa = 10.4QDD686 pKa = 3.95QPTAQTEE693 pKa = 4.24LSDD696 pKa = 3.9GLL698 pKa = 4.0
TT1 pKa = 7.16IAEE4 pKa = 4.77CLPCGKK10 pKa = 7.74QHH12 pKa = 7.23KK13 pKa = 10.58NFDD16 pKa = 4.47LDD18 pKa = 3.66PAPLIKK24 pKa = 10.42CPPGTCFRR32 pKa = 11.84QNMCIPSNSGRR43 pKa = 11.84FQCAPCPDD51 pKa = 4.17GFTGDD56 pKa = 4.38GVHH59 pKa = 7.02CDD61 pKa = 5.04DD62 pKa = 5.32VDD64 pKa = 3.52EE65 pKa = 5.54CKK67 pKa = 10.62FNPCFPGVRR76 pKa = 11.84CVNTAPGFRR85 pKa = 11.84CEE87 pKa = 4.09KK88 pKa = 10.56CPLGYY93 pKa = 9.12TGPLLTGVGVSYY105 pKa = 11.38AKK107 pKa = 9.9TNKK110 pKa = 9.05QVCEE114 pKa = 5.17DD115 pKa = 3.46IDD117 pKa = 3.98EE118 pKa = 4.7CLGPPDD124 pKa = 5.25NGGCTANSDD133 pKa = 3.94CHH135 pKa = 5.11NTMGSFRR142 pKa = 11.84CGNCKK147 pKa = 10.33SGFTGDD153 pKa = 3.73QVRR156 pKa = 11.84GCHH159 pKa = 6.9GDD161 pKa = 3.47RR162 pKa = 11.84LCPNGQPNPCDD173 pKa = 3.35INAEE177 pKa = 4.41CIVEE181 pKa = 4.0RR182 pKa = 11.84DD183 pKa = 3.56GSISCACGVGWAGNGYY199 pKa = 9.96VCGKK203 pKa = 9.08DD204 pKa = 3.26TDD206 pKa = 4.13IDD208 pKa = 3.69AHH210 pKa = 6.92PDD212 pKa = 3.21DD213 pKa = 4.71TLQCRR218 pKa = 11.84DD219 pKa = 3.77STCKK223 pKa = 9.81QDD225 pKa = 3.25NCVFVPNSGQEE236 pKa = 3.9DD237 pKa = 4.01ADD239 pKa = 4.23RR240 pKa = 11.84DD241 pKa = 4.12GMGDD245 pKa = 3.44ACDD248 pKa = 5.25DD249 pKa = 4.49DD250 pKa = 6.68ADD252 pKa = 4.25SDD254 pKa = 4.63GIMNIDD260 pKa = 3.79DD261 pKa = 4.27NCWLVPNVNQKK272 pKa = 10.44NSDD275 pKa = 3.57KK276 pKa = 10.78DD277 pKa = 3.66LHH279 pKa = 7.57GDD281 pKa = 3.17ACDD284 pKa = 3.37NCKK287 pKa = 9.07TVEE290 pKa = 4.15NPSQRR295 pKa = 11.84DD296 pKa = 3.21TDD298 pKa = 3.54QDD300 pKa = 3.95GLGDD304 pKa = 3.9EE305 pKa = 5.58CDD307 pKa = 4.71DD308 pKa = 5.76DD309 pKa = 4.25MDD311 pKa = 6.43GDD313 pKa = 4.13GLKK316 pKa = 10.81NILDD320 pKa = 3.68NCQRR324 pKa = 11.84VPNLDD329 pKa = 3.29QKK331 pKa = 11.54DD332 pKa = 3.66RR333 pKa = 11.84DD334 pKa = 3.67NDD336 pKa = 4.11GVGDD340 pKa = 4.22ACDD343 pKa = 3.93SCPDD347 pKa = 3.54MANPNQSDD355 pKa = 3.51SDD357 pKa = 4.12DD358 pKa = 4.47DD359 pKa = 4.96LVGDD363 pKa = 4.17TCDD366 pKa = 5.48DD367 pKa = 4.73NIDD370 pKa = 3.69SDD372 pKa = 4.95GDD374 pKa = 3.6GHH376 pKa = 7.04QNTKK380 pKa = 10.88DD381 pKa = 3.38NCPDD385 pKa = 3.72VINSSQLDD393 pKa = 3.62TDD395 pKa = 3.7KK396 pKa = 11.69DD397 pKa = 4.0GMGDD401 pKa = 3.49EE402 pKa = 5.68CDD404 pKa = 5.62DD405 pKa = 5.68DD406 pKa = 6.32DD407 pKa = 7.09DD408 pKa = 6.04NDD410 pKa = 5.26GILDD414 pKa = 3.81EE415 pKa = 6.04DD416 pKa = 4.7DD417 pKa = 3.7NCRR420 pKa = 11.84LVPNPDD426 pKa = 3.22QEE428 pKa = 4.67DD429 pKa = 3.77ANKK432 pKa = 10.9DD433 pKa = 3.63KK434 pKa = 11.66VGDD437 pKa = 3.7ACEE440 pKa = 4.28GDD442 pKa = 3.48FDD444 pKa = 4.85KK445 pKa = 11.65DD446 pKa = 3.6NVIDD450 pKa = 5.66IIDD453 pKa = 4.07HH454 pKa = 6.03CPEE457 pKa = 3.9NAEE460 pKa = 4.03VTLTDD465 pKa = 3.81FRR467 pKa = 11.84AYY469 pKa = 8.19QTVVLDD475 pKa = 4.24PEE477 pKa = 5.01GDD479 pKa = 3.9SQIDD483 pKa = 3.89PNWVVLNQGMEE494 pKa = 4.11IVQTMNSDD502 pKa = 3.19PGLAVGYY509 pKa = 9.83KK510 pKa = 10.02AFSGVDD516 pKa = 3.74FEE518 pKa = 5.24GTFHH522 pKa = 6.81VNTVTDD528 pKa = 3.57DD529 pKa = 4.08DD530 pKa = 4.35YY531 pKa = 12.0AGFIFGYY538 pKa = 9.58QDD540 pKa = 2.83SSSFYY545 pKa = 10.47VVMWKK550 pKa = 7.78QTEE553 pKa = 3.84QTYY556 pKa = 8.18WQAAPFRR563 pKa = 11.84AVADD567 pKa = 3.86PGIQLKK573 pKa = 10.07AVKK576 pKa = 10.09SKK578 pKa = 9.5TGPGEE583 pKa = 3.83YY584 pKa = 10.03LRR586 pKa = 11.84NSLWHH591 pKa = 6.45TGNTPDD597 pKa = 5.06QVRR600 pKa = 11.84LLWKK604 pKa = 10.3DD605 pKa = 3.22PRR607 pKa = 11.84NVGWKK612 pKa = 10.39DD613 pKa = 3.18KK614 pKa = 11.21VSYY617 pKa = 10.33RR618 pKa = 11.84WFLQHH623 pKa = 6.81RR624 pKa = 11.84PQVGYY629 pKa = 9.82IRR631 pKa = 11.84ARR633 pKa = 11.84FFEE636 pKa = 4.37GSNLVADD643 pKa = 4.25TGMITDD649 pKa = 3.77TSMRR653 pKa = 11.84GGRR656 pKa = 11.84LGVFCFSQEE665 pKa = 3.94NIIWSNLKK673 pKa = 9.68YY674 pKa = 10.31RR675 pKa = 11.84CNDD678 pKa = 4.36TIPADD683 pKa = 3.95YY684 pKa = 10.4QDD686 pKa = 3.95QPTAQTEE693 pKa = 4.24LSDD696 pKa = 3.9GLL698 pKa = 4.0
Molecular weight: 76.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q1F122|A0A3Q1F122_9TELE DENN domain containing 2B OS=Acanthochromis polyacanthus OX=80966 PE=4 SV=1
MM1 pKa = 6.95FQSSHH6 pKa = 3.95KK7 pKa = 8.31TFRR10 pKa = 11.84IKK12 pKa = 10.64RR13 pKa = 11.84FLAKK17 pKa = 9.71KK18 pKa = 9.58QKK20 pKa = 8.69QNRR23 pKa = 11.84PIPQWIRR30 pKa = 11.84MKK32 pKa = 9.89TGNKK36 pKa = 8.61IRR38 pKa = 11.84YY39 pKa = 7.09NSKK42 pKa = 8.3RR43 pKa = 11.84RR44 pKa = 11.84HH45 pKa = 3.95WRR47 pKa = 11.84RR48 pKa = 11.84TKK50 pKa = 10.83LGLL53 pKa = 3.67
MM1 pKa = 6.95FQSSHH6 pKa = 3.95KK7 pKa = 8.31TFRR10 pKa = 11.84IKK12 pKa = 10.64RR13 pKa = 11.84FLAKK17 pKa = 9.71KK18 pKa = 9.58QKK20 pKa = 8.69QNRR23 pKa = 11.84PIPQWIRR30 pKa = 11.84MKK32 pKa = 9.89TGNKK36 pKa = 8.61IRR38 pKa = 11.84YY39 pKa = 7.09NSKK42 pKa = 8.3RR43 pKa = 11.84RR44 pKa = 11.84HH45 pKa = 3.95WRR47 pKa = 11.84RR48 pKa = 11.84TKK50 pKa = 10.83LGLL53 pKa = 3.67
Molecular weight: 6.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
18433522 |
16 |
8635 |
550.6 |
61.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.528 ± 0.011 | 2.261 ± 0.011 |
5.22 ± 0.009 | 6.817 ± 0.018 |
3.636 ± 0.01 | 6.288 ± 0.015 |
2.67 ± 0.006 | 4.343 ± 0.01 |
5.603 ± 0.013 | 9.491 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.383 ± 0.006 | 3.813 ± 0.008 |
5.743 ± 0.017 | 4.771 ± 0.012 |
5.665 ± 0.012 | 8.853 ± 0.019 |
5.636 ± 0.01 | 6.336 ± 0.011 |
1.144 ± 0.005 | 2.739 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
