
Leisingera sp. ANG1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Leisingera; unclassified Leisingera
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
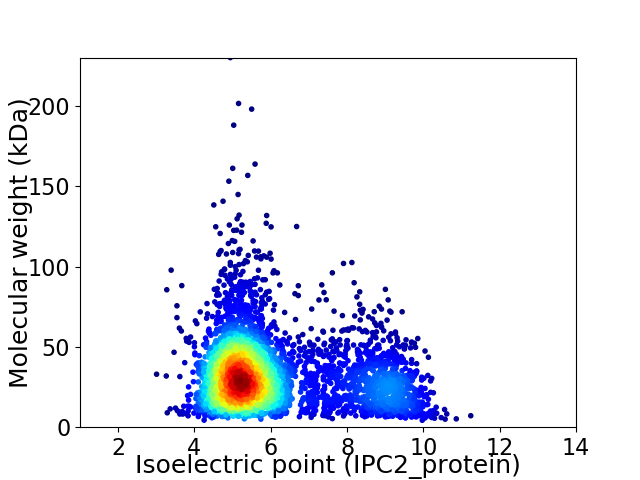
Virtual 2D-PAGE plot for 4159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4EAY3|A0A0B4EAY3_9RHOB Uncharacterized protein OS=Leisingera sp. ANG1 OX=1002340 GN=GC1_00830 PE=4 SV=1
MM1 pKa = 7.66PSGYY5 pKa = 10.19LVQLGDD11 pKa = 3.46GSLDD15 pKa = 3.4HH16 pKa = 7.36GDD18 pKa = 4.5TITSTYY24 pKa = 11.37ASFTTDD30 pKa = 2.92QNLGAGNWSWTGSHH44 pKa = 6.71GGTPYY49 pKa = 10.61TNEE52 pKa = 4.27LEE54 pKa = 4.31PGVYY58 pKa = 10.42YY59 pKa = 10.92LATDD63 pKa = 3.73GNVYY67 pKa = 9.72FVPDD71 pKa = 4.14YY72 pKa = 11.06GPPDD76 pKa = 3.81TFSSASADD84 pKa = 3.63NPPAYY89 pKa = 10.24SDD91 pKa = 3.03TDD93 pKa = 3.8GAVEE97 pKa = 4.3GTSGADD103 pKa = 3.71TIDD106 pKa = 3.53SSYY109 pKa = 11.26TDD111 pKa = 3.94ADD113 pKa = 3.56GDD115 pKa = 4.59QIDD118 pKa = 3.95NGNGGGASGNQDD130 pKa = 3.08TVEE133 pKa = 4.5AGAGDD138 pKa = 3.91DD139 pKa = 4.13TVDD142 pKa = 3.7AGADD146 pKa = 3.13NDD148 pKa = 4.04MVFGGEE154 pKa = 4.54GGDD157 pKa = 3.81SLAGGSGDD165 pKa = 3.41DD166 pKa = 4.1TIYY169 pKa = 11.25GDD171 pKa = 3.83SSSEE175 pKa = 4.01AGLTDD180 pKa = 3.52SDD182 pKa = 5.18QSITDD187 pKa = 3.67ANVTDD192 pKa = 3.95TSSGFTVTAKK202 pKa = 9.66TVAGNTSAANIDD214 pKa = 4.32YY215 pKa = 10.78YY216 pKa = 11.47DD217 pKa = 3.63GGYY220 pKa = 10.33GVAGAISDD228 pKa = 3.82SDD230 pKa = 3.92SGVDD234 pKa = 3.67AQIGYY239 pKa = 10.13DD240 pKa = 3.28KK241 pKa = 11.3ASGVSEE247 pKa = 3.94EE248 pKa = 5.17LIVDD252 pKa = 4.38FDD254 pKa = 3.67QDD256 pKa = 3.33TDD258 pKa = 3.95DD259 pKa = 3.76VSVTFKK265 pKa = 10.86HH266 pKa = 6.76LYY268 pKa = 8.03SQSNGEE274 pKa = 3.96EE275 pKa = 4.32GHH277 pKa = 5.12WAIYY281 pKa = 10.0KK282 pKa = 10.65DD283 pKa = 3.78GVLVAQGDD291 pKa = 3.91FTEE294 pKa = 5.12DD295 pKa = 3.17VAGSGTGTIDD305 pKa = 2.73ISGYY309 pKa = 11.13GDD311 pKa = 3.79FDD313 pKa = 4.28QLVLSANLQTDD324 pKa = 3.69GTDD327 pKa = 3.24GSDD330 pKa = 4.0YY331 pKa = 10.39MVTDD335 pKa = 3.41ITFAVPPEE343 pKa = 4.21VPADD347 pKa = 3.85GDD349 pKa = 3.88DD350 pKa = 4.89SIDD353 pKa = 3.94GGDD356 pKa = 4.06GDD358 pKa = 5.27DD359 pKa = 4.24VIYY362 pKa = 10.43GQGGDD367 pKa = 3.7DD368 pKa = 4.11TISGGSGADD377 pKa = 3.48TLDD380 pKa = 3.78GGAGDD385 pKa = 4.66DD386 pKa = 4.75SIIAGEE392 pKa = 4.33GDD394 pKa = 3.96SASGGAGDD402 pKa = 5.11DD403 pKa = 3.79YY404 pKa = 10.95FTIVSDD410 pKa = 3.68EE411 pKa = 4.23AFVTGDD417 pKa = 3.09ITITGGDD424 pKa = 3.79DD425 pKa = 3.83GEE427 pKa = 4.83TTGDD431 pKa = 3.38TLDD434 pKa = 4.93FNGQLDD440 pKa = 3.72KK441 pKa = 10.89GTLVITDD448 pKa = 3.71DD449 pKa = 3.75TGGSKK454 pKa = 9.21TGSATLLDD462 pKa = 3.99GSTVYY467 pKa = 10.38FSGIEE472 pKa = 3.95NIICFTAGTRR482 pKa = 11.84IATSGGLRR490 pKa = 11.84AIEE493 pKa = 3.96TLRR496 pKa = 11.84PGDD499 pKa = 3.81LVMTRR504 pKa = 11.84DD505 pKa = 3.48NGLQPVRR512 pKa = 11.84WISQSTVPATGNLAPIRR529 pKa = 11.84IKK531 pKa = 10.68AGQFGAEE538 pKa = 3.98RR539 pKa = 11.84DD540 pKa = 3.93LLVSPQHH547 pKa = 6.73RR548 pKa = 11.84MLFSGSQASLLFGEE562 pKa = 4.8SEE564 pKa = 4.43VLVSAVHH571 pKa = 5.52MLNDD575 pKa = 3.5RR576 pKa = 11.84SIRR579 pKa = 11.84RR580 pKa = 11.84EE581 pKa = 3.57PGGTVTYY588 pKa = 9.84VHH590 pKa = 6.33VLFGQHH596 pKa = 5.92EE597 pKa = 4.44IIYY600 pKa = 10.38AEE602 pKa = 4.54GAASEE607 pKa = 4.53SFHH610 pKa = 6.97PGVQAVSALEE620 pKa = 3.93EE621 pKa = 4.12PVRR624 pKa = 11.84EE625 pKa = 4.06EE626 pKa = 4.72LFCIMPDD633 pKa = 3.71LRR635 pKa = 11.84WHH637 pKa = 5.98SGEE640 pKa = 3.92GGRR643 pKa = 11.84TARR646 pKa = 11.84LCASRR651 pKa = 11.84CEE653 pKa = 4.15AALLLPPSPLDD664 pKa = 3.28QQ665 pKa = 5.04
MM1 pKa = 7.66PSGYY5 pKa = 10.19LVQLGDD11 pKa = 3.46GSLDD15 pKa = 3.4HH16 pKa = 7.36GDD18 pKa = 4.5TITSTYY24 pKa = 11.37ASFTTDD30 pKa = 2.92QNLGAGNWSWTGSHH44 pKa = 6.71GGTPYY49 pKa = 10.61TNEE52 pKa = 4.27LEE54 pKa = 4.31PGVYY58 pKa = 10.42YY59 pKa = 10.92LATDD63 pKa = 3.73GNVYY67 pKa = 9.72FVPDD71 pKa = 4.14YY72 pKa = 11.06GPPDD76 pKa = 3.81TFSSASADD84 pKa = 3.63NPPAYY89 pKa = 10.24SDD91 pKa = 3.03TDD93 pKa = 3.8GAVEE97 pKa = 4.3GTSGADD103 pKa = 3.71TIDD106 pKa = 3.53SSYY109 pKa = 11.26TDD111 pKa = 3.94ADD113 pKa = 3.56GDD115 pKa = 4.59QIDD118 pKa = 3.95NGNGGGASGNQDD130 pKa = 3.08TVEE133 pKa = 4.5AGAGDD138 pKa = 3.91DD139 pKa = 4.13TVDD142 pKa = 3.7AGADD146 pKa = 3.13NDD148 pKa = 4.04MVFGGEE154 pKa = 4.54GGDD157 pKa = 3.81SLAGGSGDD165 pKa = 3.41DD166 pKa = 4.1TIYY169 pKa = 11.25GDD171 pKa = 3.83SSSEE175 pKa = 4.01AGLTDD180 pKa = 3.52SDD182 pKa = 5.18QSITDD187 pKa = 3.67ANVTDD192 pKa = 3.95TSSGFTVTAKK202 pKa = 9.66TVAGNTSAANIDD214 pKa = 4.32YY215 pKa = 10.78YY216 pKa = 11.47DD217 pKa = 3.63GGYY220 pKa = 10.33GVAGAISDD228 pKa = 3.82SDD230 pKa = 3.92SGVDD234 pKa = 3.67AQIGYY239 pKa = 10.13DD240 pKa = 3.28KK241 pKa = 11.3ASGVSEE247 pKa = 3.94EE248 pKa = 5.17LIVDD252 pKa = 4.38FDD254 pKa = 3.67QDD256 pKa = 3.33TDD258 pKa = 3.95DD259 pKa = 3.76VSVTFKK265 pKa = 10.86HH266 pKa = 6.76LYY268 pKa = 8.03SQSNGEE274 pKa = 3.96EE275 pKa = 4.32GHH277 pKa = 5.12WAIYY281 pKa = 10.0KK282 pKa = 10.65DD283 pKa = 3.78GVLVAQGDD291 pKa = 3.91FTEE294 pKa = 5.12DD295 pKa = 3.17VAGSGTGTIDD305 pKa = 2.73ISGYY309 pKa = 11.13GDD311 pKa = 3.79FDD313 pKa = 4.28QLVLSANLQTDD324 pKa = 3.69GTDD327 pKa = 3.24GSDD330 pKa = 4.0YY331 pKa = 10.39MVTDD335 pKa = 3.41ITFAVPPEE343 pKa = 4.21VPADD347 pKa = 3.85GDD349 pKa = 3.88DD350 pKa = 4.89SIDD353 pKa = 3.94GGDD356 pKa = 4.06GDD358 pKa = 5.27DD359 pKa = 4.24VIYY362 pKa = 10.43GQGGDD367 pKa = 3.7DD368 pKa = 4.11TISGGSGADD377 pKa = 3.48TLDD380 pKa = 3.78GGAGDD385 pKa = 4.66DD386 pKa = 4.75SIIAGEE392 pKa = 4.33GDD394 pKa = 3.96SASGGAGDD402 pKa = 5.11DD403 pKa = 3.79YY404 pKa = 10.95FTIVSDD410 pKa = 3.68EE411 pKa = 4.23AFVTGDD417 pKa = 3.09ITITGGDD424 pKa = 3.79DD425 pKa = 3.83GEE427 pKa = 4.83TTGDD431 pKa = 3.38TLDD434 pKa = 4.93FNGQLDD440 pKa = 3.72KK441 pKa = 10.89GTLVITDD448 pKa = 3.71DD449 pKa = 3.75TGGSKK454 pKa = 9.21TGSATLLDD462 pKa = 3.99GSTVYY467 pKa = 10.38FSGIEE472 pKa = 3.95NIICFTAGTRR482 pKa = 11.84IATSGGLRR490 pKa = 11.84AIEE493 pKa = 3.96TLRR496 pKa = 11.84PGDD499 pKa = 3.81LVMTRR504 pKa = 11.84DD505 pKa = 3.48NGLQPVRR512 pKa = 11.84WISQSTVPATGNLAPIRR529 pKa = 11.84IKK531 pKa = 10.68AGQFGAEE538 pKa = 3.98RR539 pKa = 11.84DD540 pKa = 3.93LLVSPQHH547 pKa = 6.73RR548 pKa = 11.84MLFSGSQASLLFGEE562 pKa = 4.8SEE564 pKa = 4.43VLVSAVHH571 pKa = 5.52MLNDD575 pKa = 3.5RR576 pKa = 11.84SIRR579 pKa = 11.84RR580 pKa = 11.84EE581 pKa = 3.57PGGTVTYY588 pKa = 9.84VHH590 pKa = 6.33VLFGQHH596 pKa = 5.92EE597 pKa = 4.44IIYY600 pKa = 10.38AEE602 pKa = 4.54GAASEE607 pKa = 4.53SFHH610 pKa = 6.97PGVQAVSALEE620 pKa = 3.93EE621 pKa = 4.12PVRR624 pKa = 11.84EE625 pKa = 4.06EE626 pKa = 4.72LFCIMPDD633 pKa = 3.71LRR635 pKa = 11.84WHH637 pKa = 5.98SGEE640 pKa = 3.92GGRR643 pKa = 11.84TARR646 pKa = 11.84LCASRR651 pKa = 11.84CEE653 pKa = 4.15AALLLPPSPLDD664 pKa = 3.28QQ665 pKa = 5.04
Molecular weight: 68.25 kDa
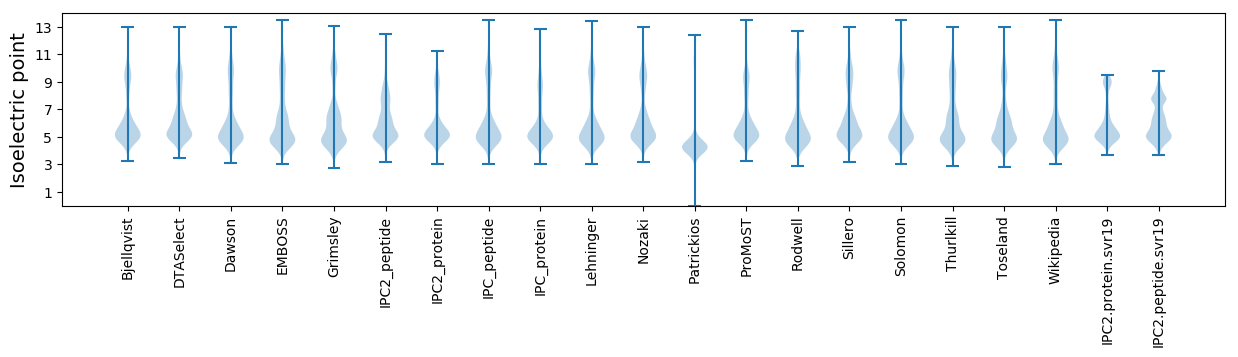
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4EHH4|A0A0B4EHH4_9RHOB Chromate transporter OS=Leisingera sp. ANG1 OX=1002340 GN=GC1_00990 PE=3 SV=1
MM1 pKa = 6.81NANWIIRR8 pKa = 11.84MVLRR12 pKa = 11.84RR13 pKa = 11.84LIGRR17 pKa = 11.84AVNAGLRR24 pKa = 11.84RR25 pKa = 11.84AAQHH29 pKa = 5.75GAQRR33 pKa = 11.84LGKK36 pKa = 10.1SKK38 pKa = 10.84GPAPGFGAGPSPQRR52 pKa = 11.84KK53 pKa = 8.27FRR55 pKa = 11.84NARR58 pKa = 11.84SRR60 pKa = 11.84IPKK63 pKa = 9.63VKK65 pKa = 10.52
MM1 pKa = 6.81NANWIIRR8 pKa = 11.84MVLRR12 pKa = 11.84RR13 pKa = 11.84LIGRR17 pKa = 11.84AVNAGLRR24 pKa = 11.84RR25 pKa = 11.84AAQHH29 pKa = 5.75GAQRR33 pKa = 11.84LGKK36 pKa = 10.1SKK38 pKa = 10.84GPAPGFGAGPSPQRR52 pKa = 11.84KK53 pKa = 8.27FRR55 pKa = 11.84NARR58 pKa = 11.84SRR60 pKa = 11.84IPKK63 pKa = 9.63VKK65 pKa = 10.52
Molecular weight: 7.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1298420 |
37 |
2149 |
312.2 |
33.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.523 ± 0.048 | 0.972 ± 0.013 |
5.599 ± 0.032 | 6.315 ± 0.031 |
3.76 ± 0.024 | 8.739 ± 0.038 |
2.087 ± 0.017 | 4.882 ± 0.025 |
3.511 ± 0.03 | 10.139 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.81 ± 0.017 | 2.653 ± 0.018 |
5.025 ± 0.028 | 3.57 ± 0.022 |
6.303 ± 0.034 | 5.269 ± 0.024 |
5.148 ± 0.025 | 6.994 ± 0.032 |
1.398 ± 0.016 | 2.303 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
